About the Administrators Guide
You can use the GenePattern public server hosted at the Broad Institute, install a local GenePattern server for your own use, or install a networked GenePattern server to be used by several people. Concepts explains the benefits of each approach.
- If you are using the GenePattern public server at http://cloud.genepattern.org/gp/, you do not need to manage the server; the GenePattern team does it for you.
- If you are installing a local GenePattern server, you will most likely use the default server settings; however, you are the GenePattern server administrator for your server and have full access to configuration options described in this guide.
- If you are installing a networked GenePattern server for use by several users, read this guide carefully. You are the GenePattern server administrator and will want to configure the server appropriately for your site.
GenePattern can be run standalone on a small machine or separated into its client and server components to take advantage of a more powerful compute server. When you install a GenePattern server, you set basic server configuration options. If you are installing a local GenePattern server for your own use, you generally do not need to modify the server configuration. If you are the server administrator for a networked GenePattern server, you generally want to modify several of the GenePattern configuration options described in this guide.
Creating Groups and Administrators
Note: Only the GenePattern team can create groups on the GenePattern public server. To create a group, you must have installed a local GenePattern server (see Starting Your Own GenePattern Server).
The GenePattern configuration file GenePatternServer/resources/userGroups.xml defines groups and group membership. The Users and Groups server settings page lists all registered users and the groups to which they belong.
To create or modify groups, edit the userGroups.xml file. The XML syntax is simple but must be followed carefully. The rules are as follows:
- Use the
<group>element to create a group. You can create any number of groups. The group names must be unique. They should include only alphanumeric characters, periods (.), and underscores (_). - Use the
<user name>element to add members to a group. You can add any number of users to a group. A user may be in any number of groups. Settinguser name = “*”adds all users to a group. - Warning: Do not delete the administrators group. GenePattern requires it.
- Anytime you make a change to userGroups.xml you will need to reload the user groups either via the Users and Groups admin page on your server or by rebooting your server. (the former is less disruptive if you are not the only user)
Creating an Administrators Group
As shown below, the default userGroups.xml file defines one group, administrators, which includes all GenePattern users. Members of the administrators group have full access to the GenePattern server and all jobs run on the server. Because all users are administrators, the default GenePattern installation has no concept of “private” data.
<!-- map of users to groups -->
<userGroups>
<group name="administrators">
<user name="*"/>
</group>
</userGroups>
To maximize data privacy, minimize the number of users in the administrators group. For example, add exactly one person to the administrators group and only that one administrator can view all jobs run on the server. Other users can view their own jobs and jobs that have been explicitly shared.
<!-- map of users to groups -->
<userGroups>
<group name="administrators">
<user name="jsmith"/>
</group>
</userGroups>
Creating Other Groups
To create a new group, add a <group> element to the userGroups.xml file. The following edited userGroups.xml file adds a second user to the administrators group and creates a new group, mjones_lab:
<!-- map of users to groups --> <userGroups> <group name="administrators"> <user name="jsmith"/> <user name="mjones"/> </group> <group name="mjones_lab"> <user name="mjones"/> <user name="jdoe"/> <user name="sfederan"/> </group> </userGroups>
Renaming a group does not update shared analysis results. Members of a group can share analysis results. If you rename a group, from old_name to new_name for example, the users in the old_name group are now in the new_name group. Analysis results that they shared however were shared with the old_name group. Each user who shared job results with the old_name group should edit the share options for the job and share the job results with the new_name group.
Modifying Server Settings
To modify the configuration of your GenePattern server, use the Server Settings page:
- Click Administration>Server Settings to display the Server Settings page.
- From the Server Settings pane, select the server setting that you want to modify. GenePattern displays a page of related server configuration options.
- Modify and save the server configuration options.
- Optionally, return to step 2 to change additional settings.
The following table summarizes the server settings. For more detail, click a link in the table.
|
Specify which clients have access to the server. |
|
|
Specify commands and qualifiers to be prepended to the command line used to invoke a module or pipeline. |
|
|
Create new server configuration options. |
|
|
Specify configuration options for the GenePattern database. |
|
|
Specify how long files remain on the server before being deleted. |
|
|
Display the log file for the GenePattern server. |
|
|
Work with a job queue that you have configured for use with a queuing system, such as the Load Sharing Facility (LSF) and the Sun Grid Engine (SGE). |
|
|
Specify the root directories for the programming languages used by GenePattern and the Java flags to be added to Java command lines executed by the server. |
|
|
If your organization has a web proxy between the GenePattern server and the internet, specify the proxy information required to access the internet. |
|
|
Specify the URL used to access the module repository and the suite repository. |
|
|
Shut down the GenePattern server. |
|
|
Broadcast a message to all users logged into the GenePattern server. |
|
|
Display the LSID of each module and pipeline installed on the GenePattern server. |
|
|
Display the account information and uploaded files of a selected user. |
|
|
Display account information for all users, including the groups to which they belong. |
|
|
Display the log file for the web server used by the GenePattern server. |
|
| Server File Paths | Enabling file paths on the GenePattern server. |
Access
Use the Access page to define which GenePattern clients have access to the GenePattern server. The localhost (127.0.0.1) computer cannot be denied access to the locally installed GenePattern server. This prevents you from inadvertently denying yourself access to the server.
Using the Access page to control which computers have access to the GenePattern server is the simplest way to secure your server. You can also control access to your server based on user authentication and user permissions, as described in Securing the Server. The Access page filters are applied before any user-specific authentication or permissions are checked. If your computer cannot access the server, you cannot access the server regardless of your username/password or permissions.
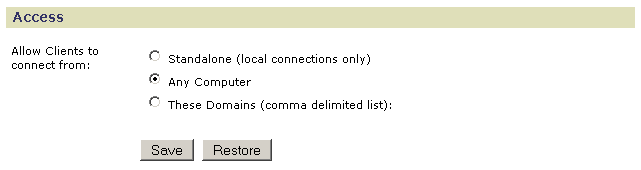
- Click Standalone to allow only local clients to connect to the server; that is, you can access this GenePattern server only from the computer that it is running on.
- Click Any Computer (default) to allow any client to connect to the server.
- Click These Domains to allow only clients from specific domains to connect to the server. Enter a comma-separated list of domains or IP addresses in the text box, for example: broadinstitute.org,dfci.harvard.edu,mit.edu.
GenePattern scans all incoming connection attempts. If they match in whole or in part any domain name or IP address in this list, the server allows access; otherwise, the server redirects the connection to a page indicating that the server does not allow access.
Click Save to save your changes. Click Restore to return to the value set at installation.
Command Line Prefix
The Command Line Prefix page allows you to prepend text to the command line used to execute a module. You can prepend the same text to all module command lines or prepend text for a specific module.
Note: Prior to GenePattern 3.2.3 (June 2010), administrators used the command line prefix for connecting to an external queuing system. GenePattern now provides the CommandExecutor interface for that purpose. For more information, see Using a Queuing System.
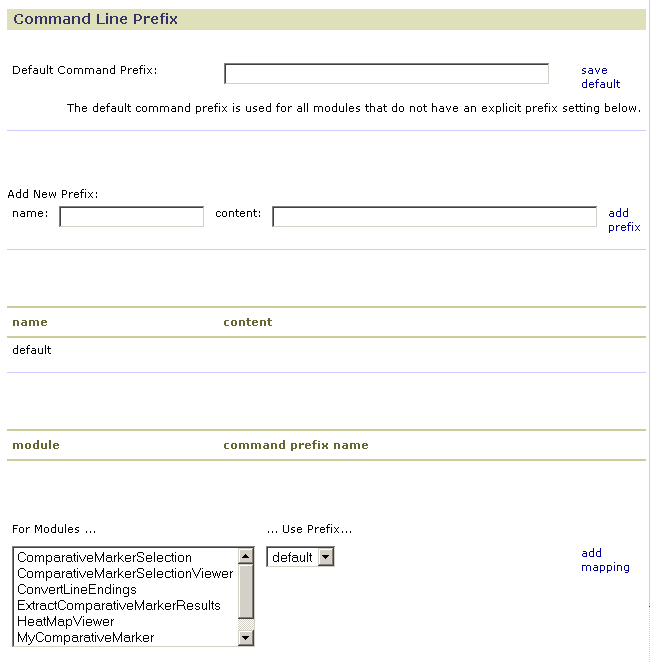
To prepend text to all (or most) command lines executed by the GenePattern server:
- Enter the desired commands and qualifiers in the Default Command Prefix field.
- Click save default. GenePattern displays the updated content of the default prefix. The name/content table in the middle of the form lists the default prefix and its content. The previous illustration shows the default prefix with no content.
When GenePattern executes a module or pipeline, it constructs the appropriate command line, prepends the default prefix to that command line, and then executes the command line.
To prepend text only to command lines that invoke specific modules or pipelines:
- In the Add New Prefix field, enter a name for the prefix and the commands and qualifiers to prepend to the command line.
- Click add prefix. GenePattern creates the new prefix, updates its content, and adds the prefix to the name/content table in the middle of the form.
- At the bottom of the form, select one or more module(s)/pipeline(s), select your new prefix, and click add mapping. GenePattern adds the prefix information to the module/command prefix name table.
When GenePattern executes a module or pipeline listed in the module/command prefix name table, it constructs the appropriate command line, prepends the specified prefix to that command line, and then executes the command line. (When GenePattern executes a module or pipeline not listed in that table, it constructs the appropriate command line, prepends the default prefix to that command line, and then executes the command line.)
Custom
Use the Custom page to define your own configuration options.
When you create a module, the custom configuration options are available as substitution variables in the module command line. For example, if you define a custom property "foo", you can use <foo> in the command line to pass the value of the custom configuration option to your module. In the Broad repository, for example, the LandmarkMatching and PeakMatch modules use the custom configuration option pepperPrefix. For more information, see Creating Modules in GenePattern.
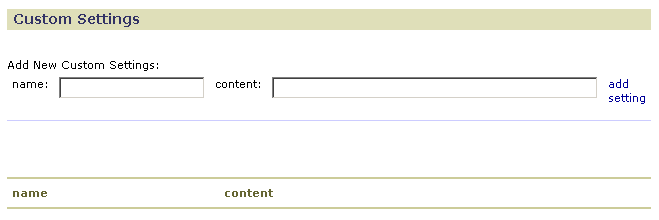
- In the name field, enter a name for the configuration option.
- In the content field, enter a value for the configuration option.
- Click add setting. GenePattern adds the option to the table at the bottom of the form.
Database
The GenePattern installation uses an in-memory HSQLDB database. You can optionally use Oracle or MySQL. To switch to a different database, create a 'database_custom.properties' file in the resources directory. The 'database_default.properties' file can be used as a reference. The 'database_example_mysql.properties' and 'database_example_oracle.properties' files are good starting points for these respective databases. Make sure to restart your server.
Starting with GenePattern version 3.9.2 the server will automatically initialize the database schema at server startup. This will work with HSQLDB, Oracle, or MySQL. For other databases you must manually initialize your schema. Use the '*.sql' files in the resources directory as a guide.
File Purge
Use the File Purge page to specify when analysis result files are deleted from the server:
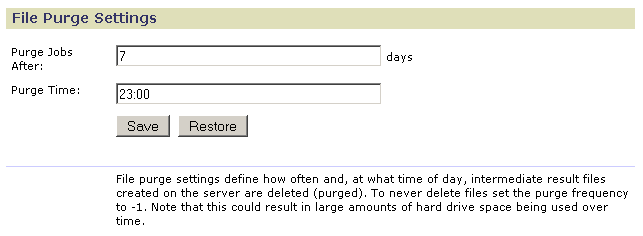
- Use Purge Jobs After to specify the number of days the server keeps the analysis result files. To prevent the server from automatically deleting the files, set this value to -1.
- Use Purge Time to specify what time of day (24-hour format) the server deletes the files.
Click Save to save your changes. Click Restore to return to the values set at installation.
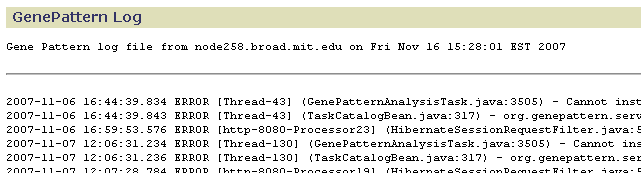
GenePattern Log
Use the GenePattern Log page to view warnings and messages generated by the GenePattern server. (Use the Web Server Log page to view messages generated by the web server that GenePattern uses.)
Locating Log Files
There are three log files that administrators might be interested in locating. These are the install log, GenePattern log and webserver log. Each of the log files can be found in the indicated location, relative to the GenePatternServer installation folder.
- Install Log: Can be located in GenePatternServer/UninstallerData/Logs.
- GenePattern Log: Can be located in GenePatternServer/logs.
- Webserver Log: Can also be located in GenePatternServer/logs.
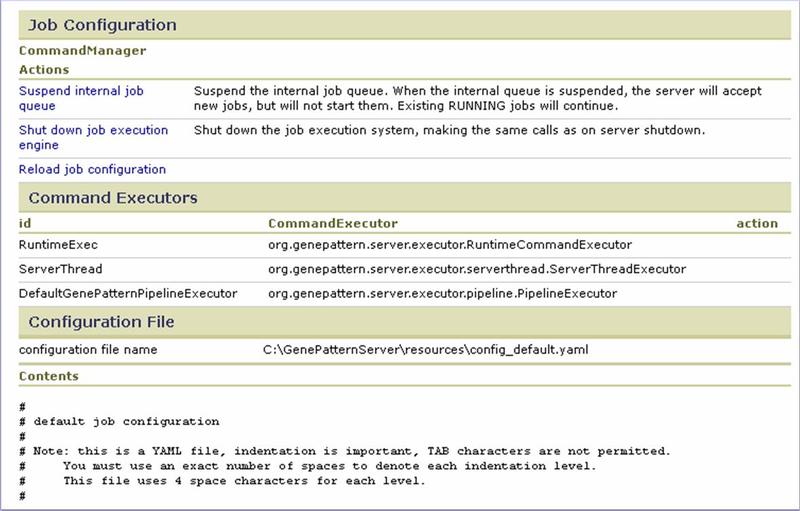
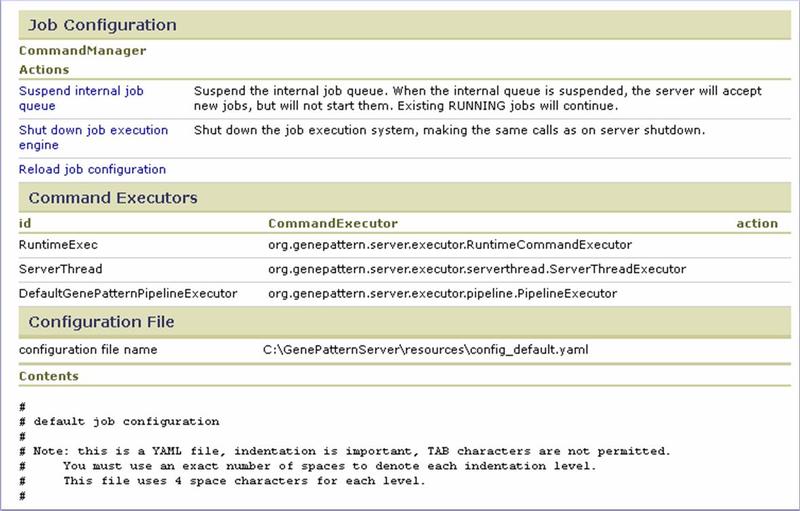
Job Configuration
If you have configured your GenePattern system to work with a queuing system, such as the Load Sharing Facility (LSF) and the Sun Grid Engine (SGE), the Job Configuration page helps you control the queue and reload your configuration files. For more information, see Using a Queuing System.
Use the Job Configuration section to control the GenePattern internal job queue:
- Suspend/Resume internal job queue. When the queuing system fails or requires maintenance, use this option to temporarily halt the internal GenePattern job queue. When the queuing system has been restarted, resume the internal GenePattern job queue.
- Shut down/Restart job execution engine. When you modify the .yaml file that configures GenePattern for use with your queuing system, you must reload that configuration file. Rather than stopping and restarting the GenePattern server, you can use this option to stop the job execution engine, click Reload job configuration to reload the configuration file, and then use this option to restart the job execution engine.
- Reload job configuration. Used in conjunction with Shut down/Restart job execution engine.
Use the Command Executors section to identify each of the command executors currently installed on the GenePattern server.
Use the Configuration File section to identify and review the .yaml configuration file currently active on the GenePattern server.
Programming Languages
The Programming Languages page contains two sections. After making changes, click Save to save them or Restore to return to the value set at installation.
Use Programming Language Configurations to specify the root directories for the programming languages used by GenePattern:
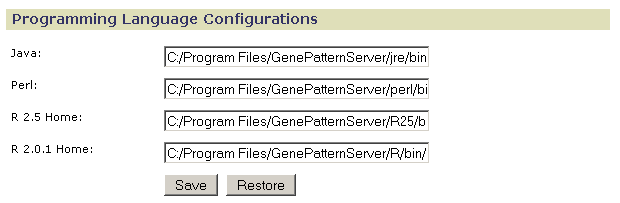
When you install GenePattern, you install the programming languages used by GenePattern. If you have alternate programming language installations that you prefer to use, use this page to point to those installations. If you would like to use more recent versions of R, see Using Different Versions of R.
Use Programming Language Options to increase the memory allocated to modules written in Java and R:
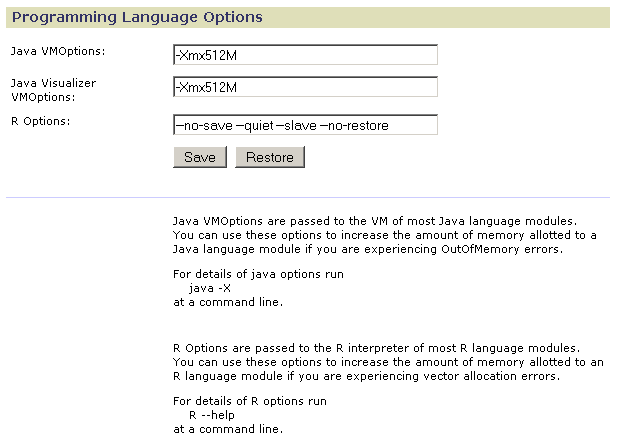
- In the Java VMOptions field, modify the Java -X flag to increase the memory allocated for Java modules run on the server (non-visualizer modules); for example, -Xmx1024M doubles the amount of memory allocated. You can specify up to the maximum memory size of your server machine. Changes take effect when you stop and restart the server.
- In the Java Visualizer VMOptions field, modify the Java -X flag to increase the memory allocated by default for modules run on the client (visualizer modules). Changes take effect when you next run a visualizer module.
GenePattern users can customize the memory allocated to visualizer modules based on the amount of memory available on their desktop PCs. To customize the memory allocated to visualizer modules run on your own desktop: select My Settings, select Visualizer Memory, and modify the Java -X flag to specify the amount of memory to be allocated to visualizer modules run on your desktop. - In the R Options field, add the --max-mem-size flag to specify the maximum amount of memory to be used by R modules running on the server; for example, --max-mem-size=1G allocates a gigabyte. You can specify up to the maximum memory size of your server machine. The --max-mem-size flag affects only Windows operating systems. Changes take effect when you stop and restart the server.
You can also increase the amount of memory allocated to the GenePattern server or client. For more information, see Increasing Memory Allocation.
Proxy
If your server is behind a firewall, use the Proxy page to set the HTTP and FTP Proxy information. Without the proxy information, the server cannot download modules, pipelines, or suites from the repository maintained by the Broad Institute. If you do not know the proxy information, contact your systems administrator.

Click Save to save your changes. Click Restore to return to the values set at installation.
Repositories
Use the Repositories page to identify the location of the repository to be accessed by the GenePattern server when you install modules and pipelines or suites from the repository. By default, it points to the module repository maintained by the Broad Institute. For information about implementing a module repository at your site, see the In-Depth Article Setting Up a Module Repository.
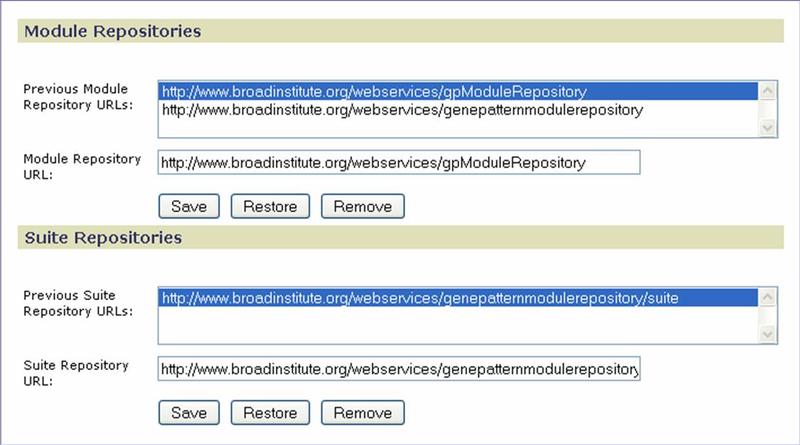
Click Save to save your changes. Click Restore to return to the values set at installation. Click Remove to delete the selected URL from the list.
Shut Down Server
You can shut down the GenePattern server by clicking the link on this page. Alternatively, double-click the Stop GenePattern Server icon on your desktop.
System Message
Use the System Message page to broadcast a message to all users logged into the GenePattern server. The message text that you enter can include simple HTML formatting commands, such as <b> and <em>.
Task Info
The Task Info page lists every module and pipeline installed on the GenePattern server. It can be useful in sorting out the confusion that can occur when modules and pipelines share the same name.
The Clear TaskInfo Cache link clears an internal GenePattern cache, which can be useful for GenePattern development. Clicking the link has no visible impact on GenePattern operations.
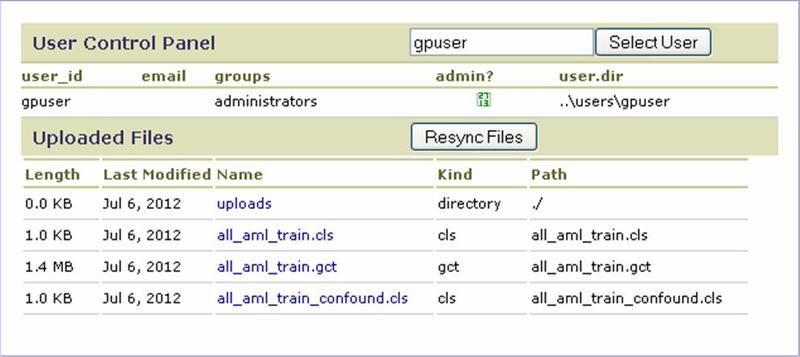
Uploaded Files
The Uploaded Files page displays basic information about a user and their uploaded files (see Uploading Files). By default, the page displays information about the user logged into the GenePattern web client. To view information for another user, enter their username and click Select User.
If a user manually adds or removes files from the Uploads directory on the file server:
- Enter their username and click Select User to display their information.
- Click Resync Files to update (resynchronize) their uploaded files based on their Uploads directory on the file server.
- Enter their username and click Select User to display the updated information.
This allows you to synchronize the user interface with the modified uploads directory on the file server without restarting the GenePattern server.
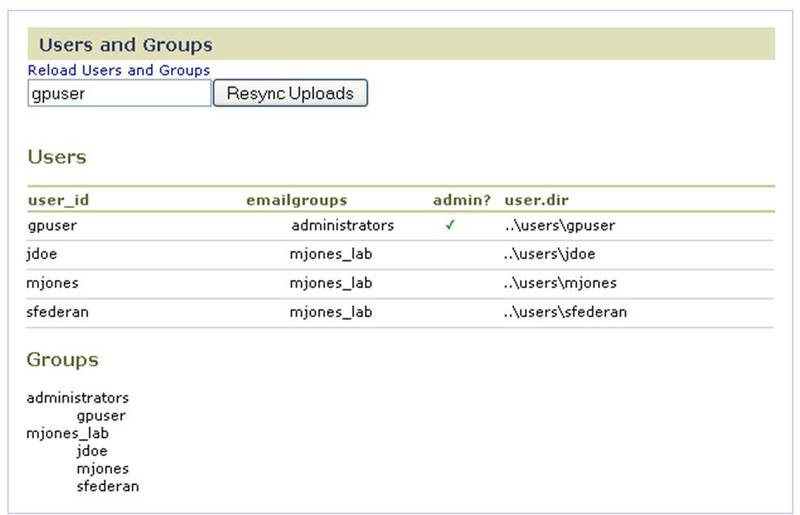
Users and Groups
Use the Users and Groups page to view user account information, including the groups to which a user belongs. This page shows only registered users. An administrator can add users to a group (Creating Groups and Administrators) before they register, but the users are not listed on this page until they have created a GenePattern account by clicking the Registration link on the GenePattern login page. If you update the userGroups.xml file, click Reload Users and Groups to update (resynchronize) the GenePattern web interface. This allows you to update users and groups without restarting the GenePattern server.
When you start the GenePattern server, the server populates the Uploads tab for each user by reviewing the Uploads directory on the file server. Typically, users add and remove uploaded files from the GenePattern web client interface. If a user adds or removes files from the Uploads directory on the file server, enter their username and click Resync Uploads to update (resynchronize) their Uploads tab based on their Uploads directory on the file server. This allows you to synchronize the user interface with the modified uploads directory on the file server without restarting the GenePattern server.
Web Server Log
Use the Web Server Log page to view messages generated by the web server that GenePattern uses. (Use the GenePattern Log page to view warnings and messages generated by the GenePattern server.)
Server File Paths
When file paths are enabled on a GenePattern server, GenePattern users can employ the Server File Paths feature to identify module and pipeline input files. With Server File Paths enabled, the GenePattern server can directly access files stored on the server's local or networked drives; therefore, input files do not have to be transferred to or stored on the GenePattern server. Avoiding file transfers can save significant upload time and avoiding file storage can save significant amounts of disk space.
Server File Paths is enabled through settings in the config_custom.yaml file (a text file in YAML format located in the GenePatternServer/resources directory). By default, the allow.input.file.paths property in the config_custom.yaml's default.properties section is set to false. Set it to true to enable Server File Paths. In addition, set the server.browse.file.system.root property to the directory where the server begins browsing for network files. Below is an example yaml entry for these two parameters:
# example config_yaml entry
default.properties:
# flag to allow users to input arbirtray network file paths as value to input file input parameters
server.browse.file.system.root: /xchip/gpbroad/shared_data
When the user clicks Add Path or URL to select a file from a local or networked drive, GenePattern opens the file selection window to /xchip/gpbroad/shared_data
A server.browse.file.system.root property definition can also reside in the YAML file's group.properties and/or user.properties sections. The group property setting will override the default property setting, and the user property setting will overide the both the default and group settings. In any of these instances (default, group, user), the server.brows.file.system.root property may be set to a list of root directories; e.g.,
server.browse.file.system.root: ["/xchip/gpbroad/shared_data", "/xchip/gpbroad/user_data/joe"]
This, for example, will lead to the display of a network file navigation tree with two roots: shared_data and joe.
Setting the Java Version
As of Release 3.9.1, the GenePattern server must be configured to run under Java 7 or Java 8. Find more details for the latest version of GenePattern in the GenePattern server release notes.
Mac
We use Oracle Java SE Development Kit (JDK) 7, and recommend you do the same. Install the JDK and not just the JRE (Java Runtime Environment).
Windows
When installed on Windows, using the installer which includes the VM, the GenePattern server is configured for Java 7 by default.
To configure the GenePattern server for Java 8:
- Confirm that Java 8 is installed.
Tip: Use the 'java -version' command. - Stop the GenePattern server.
- Edit the StartGenePatternServer.lax file and update the location of the Java executable:
# LAX.NL.CURRENT.VM
# -----------------
# the VM to use for the next launch
lax.nl.current.vm=\jre\bin\java.exe - Edit the StopGenePatternServer.lax file and update the location of the Java executable.
- Restart the GenePattern server.
Linux
When installed on Linux, the GenePattern server is configured for the version of Java in use by your Linux system by default.
To configure the GenePattern server for Java 7 or Java 8, if this is not already the case:
- Confirm that Java 7 or Java 8 is installed.
Tip: Use the 'java -version' command. - Stop the GenePattern server.
- Edit the StartGenePatternServer.lax file and update the location of the Java executable:
# LAX.NL.CURRENT.VM
# -----------------
# the VM to use for the next launch
lax.nl.current.vm=<Java>/bin/java - Edit the StopGenePatternServer.lax file and update the location of the Java executable.
- Restart the GenePattern server.
Using Different Versions of R
Installing GenePattern (version 3.1 and later) installs R 2.5.
- On Windows, R 2.5 is installed within the GenePattern directory and should not impact other installations of R that may be on your computer.
- On Mac it is installed in the standard location,
/Library/Frameworks/R.framework/Versions/2.5. - Linux users must install R on their own and point the GenePattern installation to the location of R.
Most of the GenePattern modules available in the Broad Institute repository (Modules & Pipelines>Install from repository) work with R 2.5. However, some GenePattern modules require different versions of R; for example, ComBat v2 requires R 2.7. Unfortunately, R is not backward compatible. If you simply install and run the latest version of R, modules may fail or (worse) may produce invalid results even though they do not fail. Instead, you must have multiple versions of R installed in order to run all of these modules together on the same server.
Defining R in GenePattern
In GenePattern, each module definition includes a command line that runs the analysis program. For an R module, the R version is defined by a command line substitution parameter. For example, the <R> parameter is substituted with the full path to the R 2.0.1 executable. The <R2.5> parameter is substituted with the full path to the R 2.5 executable. Similar parameters are used for other versions of R.
GenePattern version 3.1 and later installs R 2.5 and sets the <R2.5> parameter. If you upgraded from GenePattern 3.0, your GenePattern installation also includes R 2.0.1 and sets the <R> parameter.
Newer versions of GenePattern also set the <R2.5_HOME> parameter, pointing to the location of the R 2.5 installation such that Rscript is found at <R2.5_HOME>/bin/Rscript. The GenePattern team is phasing out the use of <R2.5> in favor of <R2.5_HOME> in future module revisions.
For R 2.15 and later, we are transitioning to also allow the selection of the desired Rscript executable through a similar parameter named <R2.15_Rscript>. This allows Windows users, for example, to select the 64-bit version of Rscript instead of the 32-bit executable.
We do not recommend the use R 3.0 or R 3.1 with GenePattern at this time due to the issues with compatability and validity covered earlier as we have not fully updated our components to these versions. We are actively looking at these releases for future use.
Adding More Recent Versions of R to GenePattern
To add a different version of R to your GenePattern installation (for example R 2.7 on Mac OS X, for ComBat v. 2):
- Install the required version of R, if necessary. This is covered in detail on the R Project home page. For Mac users, please read the section on 'Using Multiple Versions of R on Mac OS X' below.
- Go to http://www.r-project.org/ and select a CRAN mirror.
- Locate either the source code or the binary for the desired version of R. For a binary installation, look for the subdirectory link labeled 'old', which is towards the bottom of the page. The next section lists the locations of various versions of R used by GenePattern.
- Follow the installation instructions.
- After you install the correct version of R, in whatever manner makes sense to you, you need to configure GenePattern to use that version of R. This is as simple as adding two new substitution parameters to the server settings.
- Click Administration>Server Settings and go to the Custom page.
- Add a setting for R*_HOME and another for R*, replacing the '*' with an actual version number.
- For example, with R 2.7 on Mac OS X the parameters are:
R2.7_HOME=/Library/Frameworks/R.framework/Versions/2.7/Resources
R2.7=<java> -DR_suppress=<R.suppress.messages.file> -DR_HOME=<R2.7_HOME> -Dr_flags=<r_flags> -cp <run_r_path> RunR - For other platforms, set R2.7_HOME equal to the full path to the installation directory. It must be a directory which contains a 'bin' folder, which contains the 'R' executable.
- For example, with R 2.7 on Mac OS X the parameters are:
- For modules requiring R 2.15, it is also necessary to set the <R2.15_Rscript> parameter, pointing to the location of the 'Rscript' program on your computer. Setting the <R2.15> parameter is not required as its use is discouraged.
- On Mac OS X, the typical setting for the <R2.15_Rscript> parameter would be:
R2.15_Rscript=/Library/Frameworks/R.framework/Versions/2.15/Resources/bin/Rscript -
On Windows, the typical setting would be:
R2.15_Rscript=C:/Program Files/R/R-2.15.3/bin/x64/Rscript.exe
- On Mac OS X, the typical setting for the <R2.15_Rscript> parameter would be:
Where to Find Older Versions of R
CRAN makes older versions of R available through its archives. Archived binary releases are available here for Mac and here for Windows. Older binary releases are not available for Linux and other platforms. Instead, it is necessary to build from the archived source bundles. In particular, here are direct links to the versions of R required by modules provided in the GenePattern public and beta repositories:
- R 2.5.1: Mac, Windows, source bundle
- R 2.7.2: Mac, Windows, source bundle
- R 2.15.3: Mac, Windows, source bundle
There are a number of CRAN mirrors as well.
Using Multiple Versions of R on Mac OS X
The Simplest Path
- Install R 2.5.1 using the CRAN installer. Note that the default security settings on newer versions of Mac OS X may prevent this.
Download this patch file bundle to modify R 2.5.1 to allow use of other versions of R. After downloading, execute the following from Terminal:cd /Library/Frameworks/R.framework/Versions
sudo tar -xzvmpf ~/Downloads/R_2.5.1_mac_patch.tar.gz - (Optional) If you will be using the ComBat module, you will also need to install R 2.7.2.
Before using the CRAN installer, execute the following from Terminal:
If you do not, the CRAN installer will remove R 2.5.1. It should give you a message similar to "Forgot package 'org.r-project.R.framework' on '/'." After that, use the R 2.7.2 CRAN installer as usual.sudo pkgutil --forget org.r-project.R.framework
Download this patch file bundle to modify R 2.7.2 to allow use of other versions of R. After downloading, execute the following from Terminal:
Then set the R2.7_HOME and R2.7 substitution parameters as described above.cd /Library/Frameworks/R.framework/Versions
sudo tar -xzvmpf ~/Downloads/R_2.7.2_mac_patch.tar.gz - If you are planning to use ExpressionFileCreator v12+ or another module that requires R 2.15.3, it is recommended that you use the R Installer Plug-in which will automatically install and configure R 2.15.3 for use with GenePattern. As opportunity permits we will be updating our R 2.15.3 modules to use this plug-in, but if you encounter one which does not then you can use ExpessionFileCreator to trigger the installation.
Further Details
- For R 2.5 (and possibly earlier) to 2.10 (Tiger builds):
sudo pkgutil --forget org.r-project.R.framework - For R 2.10 to R 2.15 (Leopard builds):
sudo pkgutil --forget org.r-project.R.Leopard.fw.pkg
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/bin/R
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/bin/R32
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/bin/R64
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/etc/i386/Makeconf
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/etc/x86_64/Makeconf
- /Library/Frameworks/R.framework/Versions/<your R version>/Resources/etc/ppc/Makeconf
Adding R Version 2.0.1 to GenePattern
There are some GenePattern modules which rely on R version 2.0.1:
- CoxRegression
- LogisticRegression
- MultiplotPreprocess
- NearestTemplatePrediction
To use these modules on your server you need to add R version 2.0.1. Note that R 2.0.1 may not be available or may not work properly on newer versions of Windows and Mac OS X. We are in the process of evaluating how to address this.
To add R2.0.1 to your GenePattern installation:
- Install R2.0.1.
- In GenePattern, click Administration>Server Settings and go to the Programming Languages page.
- Set the R 2.0.1 Home parameter to the full path of the R2.0.1 installation. This defines the <R> variable.
- Click Save to update the GenePattern server configuration.
GenePattern can now run modules written for R2.0.1.
Using the R Installer Plug-in
Due to the possibly difficult nature of setting up a GenePattern server to use multiple versions of R, we have created a plug-in to assist in the process. This plug-in only deals with R 2.15 at present; we may expand to cover other versions of R in the future. The plug-in installation is triggered by the installation of a module which declares that it requires R 2.15. At the moment, this is limited to the beta releases of ExpressionFileCreator v12 and RankNormalize v1.3+ (available from our Beta repository). We will update other modules as the opportunity permits (this document last updated Feb. 27, 2015).
The Mac platform is the most tricky in terms of support for multiple versions of R and so that will be the main focus of this guide. As the story is much simpler on Windows and Linux, those platforms will be covered much more briefly.
Use of the R Installer on Mac OS X
Due to the way that it is installed, support for use of multiple versions of R on Mac OS X is tricky and has a number of issues. It is mainly due to these issues that the GenePattern team created the R Installer Plug-in to simplify the process. The plug-in will go through several possible scenarios to detect and/or install R in a way that works for GenePattern. For the most part, you should not need to worry about those details.
The Simplest Path
If you already have a version of R installed in the default location on your Mac, the plug-in will be able to set up R 2.15 for use with GenePattern. This can be any version of R: it could be 2.5.1 or 2.15.3 or any other version. The reasons for this will be discussed below if you are interested; for most users the reasons are not important.
Many users will have already installed R 2.5.1 as it required for several core GenePattern modules (CART, ComparativeMarkerSelection, ConsensusClustering, GEOImporter, NMFConsensus and others). If you think you will be using these modules, you should go ahead and install this version of R first. If you do so, then when you later install ExpressionFileCreator the plug-in will automatically download and install R 2.15.3 into the correct location and set it up for use with GenePattern.
Alternatively, if you have already installed R 2.15 (at any patch level) to the default location then the plug-in will detect it and set it up for use with GenePattern. Note that this makes some minor changes to the installed version of R. The reasons for this - and its effects - are discussed below; again, for most users the reasons are not important.
The bottom line for most users is that if you've already installed either R 2.5.1 or R 2.15 as usual from a CRAN installer, the plug-in will configure things correctly for you. Some possible complications will be discussed in the next section, but you should not need to worry about them unless you use R outside of the context of GenePattern or if you decide to update your R installation.
Possible Complications
There are some important considerations if you work with R outside of GenePattern or if you decide to update your version of R:
- On a Mac, installing a newer version of R may result in older versions being removed unless you take specific steps to avoid this. As a practical matter, the R 2.15 installers will not remove R 2.5.1.
- Certain files must be changed in the R installation in order to allow multiple versions to work at the same time. Those changes will be undone if you update R to a different patch level, so it will be necessary to re-apply those by hand. The R 2.15 patch level bundles are available from the GenePattern FTP site. After installing R 2.15.3, for example, you would download R_2.15.3_mac_patch.tar.gz and then open Terminal.app to unpack it. Execute these commands:
cd /Library/Frameworks/R.framework/Versions
sudo tar -xzvmpf ~/Downloads/R_2.15.3_mac_patch.tar.gz - If you switch between multiple versions of R outside of the context of GenePattern, you should modify all of your versions of R in a similar way. At this time, we are only providing patch level bundles for the versions of R used by GenePattern. For any other version, you will need to do this on your own. Please note that the available bundles are version-specific down to the patch level, so don't try to use them with other versions. Also, the use of a utility like RSwitch may confuse GenePattern unless all versions of R have been so modified.
- The plug-in backs up the modified files beforehand, so if you want to revert your R installation back to the original state for any reason (such as uninstalling GenePattern, for example) you can delete the modifications and put the originals back in place. Note that doing so will very likely affect the behavior of R modules in GenePattern. The original files are stored alongside the modified files in the same path location but with the ".orig" extension added (e.g. R64.orig).
- If you do not install it yourself, the version of R 2.15.3 delivered by the plug-in is not quite a full installation. In particular it is missing the R.app and R64.app GUI applications. Again, this will only those user who want to use R outside of GenePattern. These users should instead install R 2.15.3 on their own and allow the plug-in to detect it.
Using the R Installer on Windows
Support for use of multiple versions of R on a Windows machine is straightforward. R comes in standard click-through installers available from CRAN. Due to the way it is installed, each version is completely isolated and it is possible to have different major and minor R releases, and even different patch-levels, on a Windows machine at the same time: R 3.0.1, R 2.15.2, R 2.15.3, R 2.5.1, etc. can all be present with no issue. This being the case, R installation is left up to the user with the plug-in making only a few final configuration steps.
On a Windows machine, the R Installer Plug-in will attempt to detect R in the standard location. If it does, it will go ahead and configure GenePattern to use it. All you need to do is to obtain and install R 2.15.3 using the default settings, and then install a module like ExpressionFileCreator v12 that needs it.
If you will be installing R to a different location, you will need to take the extra step of setting a custom property for R2.15_HOME manually on your GenePattern server. This is also required if you decide to run a different patch level of R such as 2.15.3.
Using the R Installer on Linux
Support for use of multiple versions of R on Linux is good, though it requires platform-specific steps by the system administrator. Setting up the installations of R varies by Linux distribution; we can't give general instructions due to the large number of distributions available. You are advised to look to the specific instructions for your platform at either CRAN or your distribution's support site (or both).
Note that obtaining R through a package management system like apt-get may result in an installation that will be auto-updated to a different version in the future. This can lead to compatibility problems in running your modules and affect reproducibility of your past results. To avoid this you can instead install R from the archived source bundles to keep multiple versions available. Please refer to CRAN and your distribution's support site for more information.
After obtaining and installing the required version of R, you will need to set a custom property for R2.15_HOME manually on your GenePattern server before installing modules which require it. When the R Installer Plug-in runs, it will check whether this property has been set and verify that it points to an installation of R with the correct major and minor version.
Possible Plug-in Errors
Substitution Variable
Depending on your version of GenePattern, you may receive an error message similar to:
no substitution available R2.15_HOME in command line R2.15_HOME/bin/Rscript
Certain versions of GenePattern are not able to load all required settings in one pass but may succeed on a second attempt. Try re-installing the module or pipeline as that may resolve the issue. If this error persists, the information in this Admin Guide section and the plug-in output may help you resolve the issue. There are additional details in the Using Different Versions of R section of the Admin Guide as well.
Installation location error (Mac only)
On a Mac, if you have never installed any version of R you will probably get a message regarding the /Library/Frameworks/R.framework/Versions directory not being present. In Mac OS X Lion (10.7) and above, creating this location requires administrator privileges and can't be done within GenePattern. This is why the plug-in will work if you've already installed some version of R: that previous install will have created the correct location for you. Without that location, the installation attempt will very likely fail and require some manual steps. There are two options. Either choice is fine; you only need to do one or ther other:
- Install R 2.15 manually in the default location so that the plug-in can detect it. This is available from CRAN; see here for locations.
- Execute the following commands from a Terminal window:
sudo mkdir -p /Library/Frameworks/R.framework/Versions
sudo chgrp -R admin /Library/Frameworks/R.framework/
sudo chmod -R g+w /Library/Frameworks/R.framework/
Administrative access will be required in either case, and afterward it will be necessary to retry the plug-in installation.
Version Check
The R Installer Plug-in will check the configured version of R to make sure it is compatible. There are several problems or conditions that it can detect at this point:
- The installed R version has a different patch level than expected. This is just a warning and will not stop the plug-in installation.
- The installed R version has a different major or minor number than expected. This is considered an error; the most likely cause is that you have set up R2.15_HOME to use a version of R other than 2.15.
- The installed version of R can't execute, results in an error, or returns no output. You may need to re-install R 2.15, but first check to make sure that R2.15_HOME contains no typos.
Increasing Memory Allocation
GenePattern allocates memory to the server, to the "client" (the computer you are using to access GenePattern), and to individual modules. When a module fails with an out of memory error, you can try increasing the amount of memory allocated to the server, the client, or the module.
- Increasing Memory for Modules
- Increasing Memory for Visualizers
- Increasing Memory for the Server and/or Client
Increasing Memory for Modules
To increase the amount of memory allocated to a module written in Java or R, click Administration>Server Settings. The Programming Languages page (Programming Language Options) provides several options for increasing Java and R memory options.
You can customize memory preferences on a per-module basis. This is useful when some of your modules require more memory than others. If you haven't already done so, copy the 'config_default.yaml' file as 'config_custom.yaml'. Then set 'config.file=config_custom.yaml' in the genepattern.properties file. These files are in the resources directory of your installation. You need to restart your server for this to take effect. This ensures that your custom configuration will not be inadvertently modified when you install an updated version of GenePattern.
Set custom memory settings in the config_custom.yaml file. This is a text file in YAML format. The 'job.memory' property defines the memory requirements for your module, for example job.memory: 512m, job.memory: 2g. This defines the '-Xmx' flag passed to the java command line. It also defines the memory requirements passed along to the queuing system such as LSF or SGE. In the rare case when you want to use a different java memory flag from the queuing system flag, set both 'job.memory' and 'job.javaXmx'.
You can also customize error handling for completed jobs. When the 'job.error_status.stderr' flag is set to 'true' the server will interpret a non-empty stderr stream as a failed job. When the 'job.error_status.exit_value' flag is set to 'true' the server will interpret a non-zero exit code as a failed job.
# example config_yaml entry
module.properties:
# custom memory flags for the ConvertLineEndings module
ConvertLineEndings:
# single parameter sets both the java flag and the queuing system memory requirements
job.memory: 2Gb
#
# advanced flags
MyModule:
# it is possible to pass one value to the java command line, e.g. java -Xmx1g
job.javaXmx: 1g
# and a different value to the queuing system
job.memory: 2g
#
# ignore stderr output
job.error_status.stderr: false
# don't ignore the exit code
job.error_status.exit_value: true
job.memory=2g job.error_status.stderr=false job.error_status.exit_value=true
Increasing Memory for Visualizers
Many GenePattern modules are run on the server. However, visualizers are applications that run on your computer, rather than on the GenePattern server. This means that you must have Java installed on your computer. For easy debugging, set your Java preferences so that the Java console displays.
The default visualizer memory limit is 512 MB. However, if you find that your visualizer repeatedly runs out of memory, you can try a few things to eliminate that error:
- Increase the visualizer memory limit in My Settings in GenePattern. (Note: In GenePattern 3.2.1, the My Settings options is ignored due to a bug. This bug is corrected in GenePattern 3.2.3 and later.)
- If you are an administrator and want to make this a global change, go to the genepattern.properties file and set the visualizer_java_flags to a higher value. For 32-bit Windows machines, this will reach a maximum of ~2 GB, but Macintosh, Unix, and 64-bit Windows machines should allow you to make use of a greater portion of your machine's available memory.
- Reduce the size of the data you are attempting to load into the viewer.
Increasing Memory for the Server and/or Client
To increase the amount of memory allocated to the server and/or the client, follow the instructions for your platform:
Mac OS X
- Right-click on the file
GenePattern/Tomcat/StartGenePatternServer(server) or the GenePatternClient/GenePattern Client (client). - Select Show Package Contents from the pop-up menu. The Contents directory should open in the finder.
- In the Contents directory, double-click the
Info.plistfile. This should open the Property List Editor program. - Add the child
VMOptionsunder theJavanode. - Change the Class of the added
VMOptionsnode to ‘Array’. - Add the child with Class 'String' with the value
-Xmx512M. You can replace the value 512 with the maximum amount of memory in MB that you want the GenePattern Client to use.
Windows and Linux
- Edit the configuration file
GenePatternServer/StartGenePatternServer.lax(server) orGenePatternClient/GenePattern Client.lax(client). - In either file, look for the entries noted below and increase these values (for example, double the value) up to the maximum memory size of the machine you are using. (Note: Windows limits the total space available to a process to 2 GB. Some of that is used for overhead, so slightly less is really available to the JRE.)
lax.nl.java.option.java.heap.size.initiallax.nl.java.option.java.heap.size.max
Using a Queuing System
Queuing systems such as the Load Sharing Facility (LSF) and the Sun Grid Engine (SGE) allow computational resources to be used effectively. If you have installed a queuing system, you can configure the GenePattern server to use it. On a heavily used server, using a queuing system to execute analysis jobs generally improves performance overall, especially for compute-intensive and long-running jobs; however, short jobs might take slightly longer because they must be dispatched to the queuing system.
The GenePattern server includes support for Sun Grid Engine (SGE) and LSF. To configure your server, you need to edit the configuration file and restart the server. Detailed documentation is in the 'config_example.yaml' file which is in the resources directory of your local GenePattern installation.
There are three additional ways to configure GenePattern's interaction with your queuing system; either programmatically or with a command line prefix.
- JobRunner API. We strongly recommend (and use) this method. Although it requires a knowledge of Java, this method is more robust than the command line prefix option and simpler to use than the Command Executor Interface. The GenePattern runtime takes care of polling your system for job completion. You must provide hooks for starting a job, cancelling a job, and getting the current status of a running job.
- CommandExecutor Interface. This API also requires that you write a Java class. Your code is responsible for polling the queuing system and notifying the GenePattern server on job completion or cancellation. The GenePattern runtime has a callback method to call when a job completes.
- Command Line Prefix. Before the 3.2.3 release of GenePattern (June 2010), the command line prefix was the only way to connect to an external queuing system. Although this option requires no Java programming and allows for configuration via a web page, it has significant drawbacks. See Command Line Prefix for details.
JobRunner API
To integrate your queuing system with GenePattern:
- Implement the JobRunner API
- Deploy the jar file to the GenePattern server.
- Configure your server.
- Reload the new configuration.
1. Implement the JobRunner API
A source code snippet from the API is included here. Contact us for the full source code.
interface JobRunner {
/**
* The GenePattern Server calls this when it is ready to submit the job to the queue.
* Submit the job to the queue and return immediately.
* The drm jobId returned by this method is used as the key into a
* lookup table mapping the gp jobId to the drm jobId.
*
* @return the drm jobId resulting from adding the job to the queue.
*/
String startJob(DrmJobSubmission drmJobSubmission) throws CommandExecutorException;
/**
* Get the status of the job.
* @param drmJobId
* @return
*/
DrmJobStatus getStatus(DrmJobRecord drmJobRecord);
/**
* This method is called when the GP server wants to cancel a job before it
* has completed on the queuing system.
* For example when a user terminates a job from the web ui.
*
* @param drmJobRecord, contains a record of the job
* @return true if the job was successfully cancelled, false otherwise.
* @throws Exception
*/
boolean cancelJob(DrmJobRecord drmJobRecord) throws Exception;
}
The required Java libraries come with your local install of GenePattern and can be found in the <GenePatternServer>/Tomcat/webapps/gp/WEB-INF/lib directory.
Follow steps 2-4 in the CommandExecutor Interface section below to configure your server.
CommandExecutor Interface
To use a queuing system with GenePattern:
- Implement the CommandExecutor Java API.
- Deploy the resulting jar file to the GenePattern server.
- Configure your server.
- Reload the new configuration.
Each step is described in detail below.
1. Implement the Command Executor Interface
The full source for the Command Executor API is included here:
/**
* Interface for managing job execution via runtime exec or an external queuing system. This interface is responsible for both initialization and shutdown of external services,
* as well as the management of job submission, getting job status, and killing, pausing, and resuming jobs.
*
* @author pcarr
*/
public interface CommandExecutor {
//configuration support
/**
* [optionally] set a path to a configuration file.
*/
void setConfigurationFilename(String filename);
/**
* [optionally] provide properties.
* @param properties
*/
void setConfigurationProperties(CommandProperties properties);
/**
* Start the service, typically called at application startup.
*/
public void start();
/**
* Stop the service, typically called just before application shutdown.
*/
public void stop();
/**
* Request the service to run a GenePattern job. It is up to the service to monitor for job completion and callback to GenePattern when the job is completed.
*
* @see GenePatternAnalysisTask#handleJobCompletion(int, String, String, int)
*
* @param commandLine
* @param environmentVariables
* @param runDir
* @param stdoutFile
* @param stderrFile
* @param jobInfo
* @param stdin
*
* @throws CommandExecutorException when errors occur attempting to submit the job
*/
void runCommand(
String commandLine[],
Map<String, String> environmentVariables,
File runDir,
File stdoutFile,
File stderrFile,
JobInfo jobInfo,
File stdinFile)
throws CommandExecutorException;
/**
* Request the service to terminate a GenePattern job which is running via this service.
* @param jobInfo
* @throws Exception indicating that the job was not properly terminated.
*/
void terminateJob(JobInfo jobInfo) throws Exception;
/**
* This method is called on server startup for each RUNNING job for this queue.
*
* For RuntimeExec, tell the GP server to delete the job results directory and requeue the job.
* For other executors, (such as LSF), you may want to ignore this message.
* For PipelineExec, you may need to determine the last successfully completed step before resuming the pipeline.
*
* @return an optional int flag to update the JOB_STATUS_ID in the GP database, ignore if it is less than zero
*/
int handleRunningJob(JobInfo jobInfo) throws Exception;
}
The required Java libraries come with your local install of GenePattern and can be found in the <GenePatternServer>/Tomcat/webapps/gp/WEB-INF/lib directory.
The interface accepts requests to start and terminate jobs from the server. You will need to invoke a callback to the GP server when your job has completed.
Example snippet:
try{GenePatternAnalysisTask.handleJobCompletion(jobInfo.getJobNumber(), exitCode,null, runDir, stdoutFile, stderrFile);}catch(Exception e) {log.error("Error handling job completion for job "+jobInfo.getJobNumber(), e);}
Once you have implemented this interface, create a jar file to deploy to the GP server.
2. Deploy the jar File to the GenePattern Server
The jar file and all of the dependent libraries must be installed to <GenePatternServer>/Tomcat/webapps/gp/WEB-INF/lib.
3. Configure Your Server
To configure your server to interact with your queuing system, you must edit the config.yaml file. In a fresh install of GenePattern, there will be two .yaml files found in <GenePatternServer>/resources: config_default.yaml and config_example.yaml. It is highly recommended that you make a copy of config_default.yaml and name it something like config.yaml. This will give you a working copy of your configuration file, preserving the default and example versions for your future reference. Additionally this will prevent your working copy from getting overwritten during server upgrade.
Edit the config.file property in the <GenePatternServer>/resources/genepattern.properties file to point to your new configuration file. By default, the property looks like this:
config.file=config_default.yaml
For this example, you would edit the property as follows:
config.file=config.yaml
Now, edit your working copy of the configuration file, config.yaml. (The following code snippets come from the config_example.yaml file.)
a) Define an executor in the "executors" section. To do so add an item to the list of 'executors' in the yaml document.
# a list of command executors# The executor id,'org.genepattern.server.executor.PipelineExecutor', is reservedforthedefaultexecutor which runs all GP pipelines.# Don't usethisas an executor id inthisfile.# a map of <id>:<obj>, where# obj := <classname> | <map># classname := fully qualified classname of aclasswhichimplementsthe org.genepattern.server.executor.CommandExecutorinterface# map := classname=<classname> [configuration.file: <path_to_config_file> | configuration.properties: <map>] [default.properties: <map>]executors:#defaultexecutorforall jobs, it is included in GenePatternRuntimeExec:classname: org.genepattern.server.executor.RuntimeCommandExecutorconfiguration.properties:# the total number of jobs to run concurrentlynum.threads:20# the total number of jobs to keep on the queue, not yet implemented#max.pending.jobs:20000# nested declaration with configuration file, <id>: { classname: <classname>, configuration: <config_file> }Test:classname: org.genepattern.server.executor.TestCommandExecutorconfiguration.properties:num.threads:20
b) Configure your server to use your executor.
# apply these properties to all jobsdefault.properties:executor: Testjava_flags: -Xmx512m
c) Optionally, you can use the configuration file to override the default executor on a per module, group or user basis. The following example comes from per module section, more examples can be found in config_example.yaml.
# overridedefault.properties and executor->default.properties based on taskname or lsid# Note: executor->configuration.properties are intended to be applied at startup and are not overwritten heremodule.properties:CBS:executor: LSFlsf.max.memory:16java_flags: -Xmx16g
About the .yaml configuration file: As of GenePattern 3.4.0, you use the .yaml configuration file only to configure GenePattern for use with a queuing system. As you work with the .yaml file, you may notice that it contains several properties that are also defined in the genepattern.properties file. To avoid confusion, leave them set to agree with the genepattern.properties file. GenePattern 3.4.0 reads these properties from the genepattern.properties file, not from the .yaml file. (In a future release, the genepattern.properties file may define the default server settings and the .yaml configuration file may define custom server settings.)
4. Reload the Configuration
At this point, you have deployed your command executor, modified the .yaml configuration file to control its use, and modified the <GenePatternServer>/resources/genepattern.properties file to point to the modified .yaml configuration file. Now, stop and restart the GenePattern server to reload the server configuration and begin to use the new command executor.
As you use GenePattern with the queuing system, you may find it useful to modify the configuration. The Administration>Server Settings>Job Configuration page provides several useful tools for controlling the internal GenePattern job queue and reloading the .yaml configuration file. Use this page to confirm which command executors are currently installed and the exact .yaml configuration file currently in use. If you make minor adjustments to the configuration file, such as overriding the command executor used for a module, group or user, you can use the Job Configuration page to reload the configuration file without restarting the GenePattern server. On the other hand, for major changes, such as adding a new command executor, we recommend restarting the server rather than simply reloading the configuration.
Command Line Prefix
Pros and Cons
Before the 3.2.3 release of GenePattern (June 2010), the only way to connect to an external queuing system was to use the command line prefix. Although this option requires no Java programming and allows for configuration via a web page, it has significant drawbacks:
- Not suitable for high volume of jobs.
- Not suitable for long running jobs.
- User jobs may be terminated and not restarted.
The drawbacks are a result of how the command line prefix works. Each new job requires a dedicated server process which waits for the job to complete. When a user terminates a job, the server process is terminated but the external process launched on the queuing system is not terminated. Similarly, when the GenePattern server shuts down, all server processes halt but the processes running on the external queuing system become orphaned. When the GenePattern server restarts, the jobs are not restarted; the user must restart any unfinished job from the beginning.
If you are using the CommandExecutor Interface, we recommend that you not use the command line prefix. The command line prefix is appended to the module command line before the job is executed by the CommandExecutor. To be more precise:
- The server gets the initial command line from the manifest.
- The command line prefix is appended to the initial command line.
- The server resolves all system and user variables and breaks the command line string into tokens.
- The server calls the CommandExecutor.runCommand.
Using the Command Line Prefix
Although this is not the preferred method, you can still use the Command Line Prefix to connect to an external queuing system.
To use the Command Line Prefix to configure the GenePattern server to execute jobs using LSF or SGE:
- Add the GenePatternURL property to the GenePattern configuration file,
GenePatternServer/resources/genepattern.properties, specifying the URL of your server. For example:GenePatternURL=http://myserver.company.com:8080/gp/When you run a pipeline, the GenePattern server uses this URL to construct the links to the output files.
By default, the GenePatternURL property is not set. When you run a pipeline, the GenePattern server derives the URL at run time based on the current IP address of the host server. This is ideal for a user running on a laptop, where the IP address may change at startup. However, if you are using a queuing system, the derived URL is incorrect: it is based on the IP address of the queuing system server rather than the GenePattern server.
- For Sun Grid Engine modify the R2.5 property in the GenePattern configuration file,
GenePatternServer/resources/genepattern.properties, to quote the <r_flags> options. For example:R2.5=<java> -DR_suppress\=<R.suppress.messages.file> -DR_HOME\=<R2.5_HOME>
-Dr_flags\=\"<r_flags>\" -cp <run_r_path> RunRModify other similar properties (if any) that were added to support additional versions of R.
- Click Administration>Server Settings and use the Command Line Prefix page to have the GenePattern server add the required options to the command line each time it executes a module.
For example, if you are using LSF, modify the Command Line Prefix options as follows:
- Click Administration>Server Settings and select Command Line Prefix. GenePattern displays the Command Line Prefix page.
- Enter the following text in the Default Command Prefix field:
bsub -K -o lsf_log.txt- The –K flag instructs the bsub command to wait for the job to complete before returning.
- The –o flag specifies the file to which the job writes standard output and standard error messages.
- Optionally, set the environment variables BSUB_QUIET and BSUB_QUIET2 to prevent bsub from printing common job messages to standard out:
- Setting BSUB_QUIET prevents bsub from printing the messages <<Job is submitted to default queue <normal>>> and <<Waiting for dispatch>>.
- Setting BSUB_QUIET2 prevents bsub from printing the message <<Job is finished>>.
Another alternative is to create a script that sets the environment variables and then executes the job using LSF or SGE. The command prefix would then execute the script. For example:
- Create the shell script to set the variables and execute the job using LSF. The script executes in the jobResults directory for the job; for example, for job 3248, the script executes in the GenePattern /Tomcat/webapps/gp/jobResults/3248/ directory. The following script sets the environment variables, submits the job to the LSF queue, waits for the job to complete, saves stdout to a new file, stdout.txt, and saves stderr to a new file, stderr.txt. By convention, GenePattern considers a job to fail if there is any output to stderr.
#!/bin/bash # # Submit the job to LSF # Save lsf out and err files in the jobResults directory. # If there is stdout from the job, pipe to stdout of this script. # If there is stderr from the job, pipe to stderr of this script. lsf_err=.lsf.err; cmd_out=cmd.out; BSUB_QUIET= BSUB_QUIET2= export BSUB_QUIET export BSUB_QUIET2 # submit the job and wait (-K) for the job to complete bsub -q genepattern -K -o .lsf_%J.out -e $lsf_err $"$@" \>$cmd_out # sleep to allow for NFS delay sleep 2; # If there is stdout from the job, pipe to stdout of this script, then delete the output file if [ -e $cmd_out ] then cat $cmd_out >&1; rm $cmd_out; fi # If there is stderr from the job, pipe to stderr of this script then delete stderr file if [ -e $lsf_err ] then cat $lsf_err >&2; rm $lsf_err; fi - Click Administration>Server Settings and select Command Line Prefix. GenePattern displays the Command Line Prefix page.
- Enter the following text in the Default Command Prefix field:
/fully/qualified/path/to/lsf_default.sh - The script shown here saves the lsf log file into the job results directory. In GenePattern, the log files are displayed with the other job result files. If you do not want the log files displayed in GenePattern, edit the /resources/genepattern.properties file and set the following property:
jobs.FilenameFilter=.lsf*
Securing the Server
Secure the GenePattern server to control who has access to which operations. Since GenePattern is primarily a web application (including SOAP interfaces) running on a web server, general approaches for securing web servers are applicable to the GenePattern server. In addition, GenePattern provides several security features that can easily be used by non-technical users to control access to the server.
This section describes several ways to secure the GenePattern server:
- Access Filtering
- Password Protection
- User Accounts
- User Permissions
- User Authentication and Authorization
- Secure Sockets Layer (SSL) Support
Access Filtering
Use the Access page to define which GenePattern clients have access to the GenePattern server. This is the simplest way to secure your GenePattern server.
Access filtering prevents users from connecting to the GenePattern server unless they come from a known computer. If your computer cannot access the server, you cannot access the server regardless of your username/password or permissions. The localhost (127.0.0.1) computer cannot be denied access to the locally installed GenePattern server. This prevents you from inadvertently denying yourself access to the server.
To use access filtering (as described in Modifying Server Settings):
- Click Administration>Server Settings.
- Use the Access page to determine which clients have access to your GenePattern server:

- Click Standalone to allow only local clients to connect to the server; that is, you can access this GenePattern server only from the computer that it is running on.
- Click Any Computer (default) to allow any client to connect to the server.
- Click These Domains to allow only clients from specific domains to connect to the server. Enter a comma-separated list of domains or IP addresses in the text box, for example:
broadinstitute.org,dfci.harvard.edu,mit.edu.
GenePattern scans all incoming connection attempts. If they match in whole or in part any domain name or IP address in this list, the server allows access; otherwise, the server redirects the connection to a page indicating that the server does not allow access.
Password Protection
By default, the GenePattern server requires only a user name to authenticate a GenePattern user. You can easily add password protection by modifying the GenePattern server properties.
To add password protection, modify the GenePattern server properties:
- Edit the GenePattern configuration file,
GenePatternServer/resources/genepattern.properties. - Set the requirePassword property to true: requirePassword=true.
- Save the
genepattern.propertiesfile. - Restart the GenePattern server.
When you add password protection to the server:
- Existing users are assigned a blank password. The first time a user signs in with a blank password, the GenePattern server requires the user to set the password.
- New users are required to register before using the GenePattern server. By registering, the user creates a GenePattern account with an associated username, password, and email address.
- The sign-in screen prompts you for a username and password. If you forget your password, click the Forgot your password? link and GenePattern emails you a temporary password.
Assigning passwords to existing user accounts prevents anyone from inadvertently or intentionally logging into and taking control of another user’s account. After adding password protection to the server, set passwords for existing users as follows:
- Select Administration>Server Settings>Users and Groups to list all users registered on the server.
- Sign into GenePattern using the name of an existing user.
- When GenePattern prompts you to set a password, select a password for that user.
- After setting the password, GenePattern displays the Change Email page (My Settings). Set the user’s email address if it has not been set. This is the address GenePattern uses to send the user a new password if necessary.
- Sign out and repeat the process for the next user.
- After setting passwords for all users, let them know that passwords have been set. You do not need to send the users their passwords. Simply ask users to sign into GenePattern and click the Forgot your password? link to have GenePattern send a temporary password.
Anonymous (Un-authenticated) Login
If desired, the GenePattern server can be configured to allow users to run analyses without a password, or even registring a username or email. This is typically necessary only when athe server is part of a journal submission to an academic journal that requires free and anonymous access (e.g. anything from the Oxford University Press which states "Web services must not require mandatory registration by the user. ").
To enable this feature, add the following property into the default_properties section of your server's config_custom.yaml file:
anonymous.users.allowed: true
When this value is present, you will see an additional option to 'sign on anonymously' on the login screen to permit anonymous login and use as shown below.
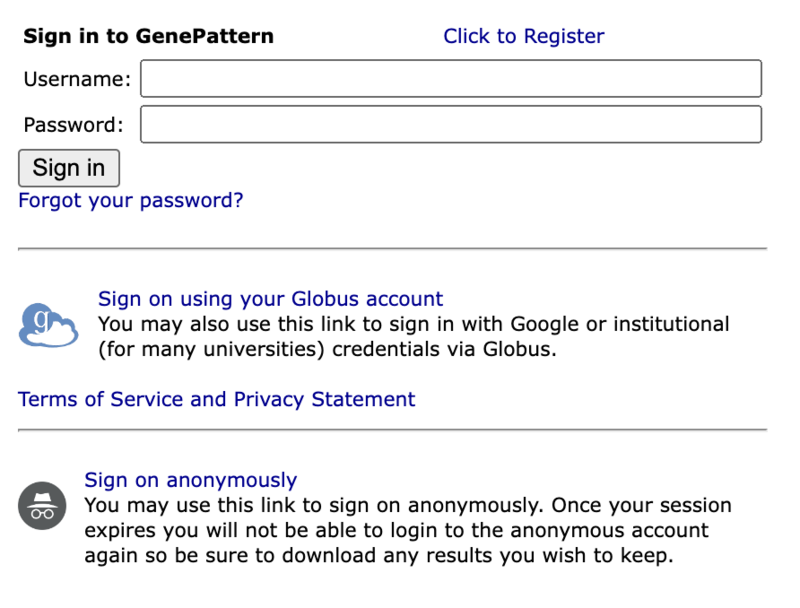
Globus Login
It is also possible to use Globus authentication to identify users for your GenePattern server. To do this you must first register your GenePattern server as an "App" with Globus authentication following these instructions.
On the GenePattern server, you will have to add the following additional configuration items to the default_properties section of your server's config_custom.yaml:
###############################
#
# Globus OAuth config settings
#
##############################
oauth.authorize.url: "https://auth.globus.org/v2/oauth2/authorize"
oauth.client.id: <your Globus oauth client id>
oauth.client.secret: <your Globus oauth secret>
oauth.client.scopes: "urn:globus:auth:scope:transfer.api.globus.org:all urn:globus:auth:scope:auth.globus.org:view_identities openid profile email offline_access"
authentication.class: org.genepattern.server.webapp.rest.api.v1.oauth.GlobusAuthentication
With this configuration and registration complete, your login page will have an additional option, "Sign on using your Globus account", shown for logging in via Globus as shown below:

User Accounts
By default, users create their own accounts by clicking the Registration link on the GenePattern login page. To configure GenePattern to allow only administrators to create new accounts:
- Shut down the server.
- Edit the file
GenePatternServer/Tomcat/webapps/gp/WEB-INF/web.xml. RemoveregisterUser.jsffrom theno.login.required.redirect.to.homeparameter value. After the edits, it looks like this:<init-param> <!-- List of jsf pages that user can access if not logged in. If user requests one of these pages while logged in, he is redirected to the home page. --> <param-name>no.login.required.redirect.to.home</param-name> <param-value>login.jsf,forgotPassword.jsf</param-value> </init-param>Result: A user cannot access the registration page until she has successfully logged into the server. - Edit the file
GenePatternServer/resources/actionPermissionMap.xml. Add the following line to the <actionPermissionMap>:
Result: A user must be an administrator to access the registration page.<url link="registerUser.jsf" permission="adminServer"/> - Edit the file
GenePatternServer/Tomcat/webapps/gp/pages/login.xhtml. Replace the phrase
withrendered="#{loginBean.createAccountAllowed and loginBean.showRegistrationLink}">
Result: Removes the Click to register link from the login page.rendered="false"> - Restart the server.
To create an account:
- Login using an administrator account. GenePattern displays the home page.
- Open the user registration page, registerUser.jsf. For example, if the URL for the GenePattern home page is
change the URL tohttp://127.0.0.1:8080/gp/pages/index.jsfhttp://127.0.0.1:8080/gp/pages/registerUser.jsf - Create the new user account. You are automatically logged in as that new user.
- To create another new account, logout of the new user account and login using your administrator account.
User Permissions
User permissions determine valid actions for the user. Permissions are based on two configuration files in the GenePatternServer/resources directory (the links show the default files):
- userGroups.xml defines user groups, as described in Creating Groups and Administrators
- permissionMap.xml defines which user groups have which permissions
A user who belongs to multiple groups is given the most permissive permissions granted to those groups. For example, an administrator who belongs to other groups retains administrator permissions.
To assign or modify user permissions, edit the permissionMap.xml file. The XML syntax is simple but must be followed carefully. The rules are as follows:
- To assign permissions to a group, add a
<group>element to that permission. A<permission>element may have any number of<group>elements. A<group>element may be listed under any number of<permission>elements. - To assign a permission to all groups (and therefore all users), use the syntax
<group name="*"/>.The presence of a group named * means that all groups (and therefore all users) have that permission. - Warning: Do not add or remove
<permission>elements. GenePattern uses them to define the permissions that it requires and implements. The permissions are described in the following table.
By default:
- When you install GenePattern, all groups have the following permissions: createPrivatePipeline, createPublicPipeline, createPrivateSuite, createPublicSuite. Administrators have all permissions.
- On the GenePattern public server, all groups have the following permissions: createPrivatePipeline, createPrivateSuite. Administrators (the GenePattern team) have all permissions.
|
Note: No explicit permission is required to run public modules/pipelines, or private modules/pipelines that you have created. No explicit permission is required to edit or delete your own modules, pipelines, suites, or jobs. |
|
|
createModule |
Permits creation of a module. Creation refers to any action that adds a module to the server, including create, install from repository, install from zip, and clone. |
|
createPrivatePipeline |
Permits creation of a private pipeline (a pipeline visible only to its creator). Creation refers to any action that adds a private pipeline to the server, including create, install from repository, install from zip, and clone. Note: To install the modules in a pipeline, you must have createModule permission. |
|
createPrivateSuite |
Permits creation of a private suite (a suite visible only to its creator). Creation refers to any action that adds a private suite to the server, including create, install from repository, install from zip, and clone. Note: To install the modules in a suite, you must have createModule permission. |
|
createPublicPipeline |
Permits creation of a public pipeline. Creation refers to any action that adds a public pipeline to the server, including create, install from repository, install from zip, and clone. Note: To install the modules in a pipeline, you must have createModule permission. |
|
createPublicSuite |
Permits creation of a public suite. Creation refers to any action that adds a public suite to the server, including create, install from repository, install from zip, and clone. Note: To install the modules in a suite, you must have createModule permission. |
|
adminJobs |
Permits viewing and deleting jobs and associated files owned by other users. Users with this permission can delete any job on the server. Typically, only members of the Administrators group are given this permission. |
|
adminModules |
Permits viewing and deleting private modules owned by other users. Permits deleting public modules. Note: No explicit permission is required to view public modules. |
|
adminPipelines |
Permits viewing and deleting private pipelines owned by other users. Permits deleting public pipelines. Note: No explicit permission is required to view public pipelines. |
|
adminSuites |
Permits viewing and deleting private suites owned by other users. Permits deleting public suites. Note: No explicit permission is required to view public suites. |
|
adminServer |
Permits access to Administration>Server Settings and all actions on the Server Settings page, including modifying server settings and shutting down the server. Users with this permission are considered to be GenePattern administrators. On the Users and Groups page, a checkmark in the admin? column indicates that a user has this permission. Typically, only members of the Administrators group are given this permission. |
User Authentication and Authorization
You can configure the GenePattern server to provide password protection, restrict creation of user accounts, and assign permissions based on groups. Additional or alternative authentication and authorization mechanisms can be added to the server by an administrator with programming experience. The remainder of this section is written for such a programmer. Note: The links in this section display the source code for the default GenePattern installation, which should be used as the starting point for any modifications.
Authentication
The authentication filter, AuthenticationFilter.java, controls whether a user can log into the server (typically based on username and password). The easiest way to modify GenePattern authentication is by implementing the IAuthenticationPlugin.java interface:
- Implement the
IAuthenticationPlugininterface. Use the IAuthenticationPlugin.java file as the starting point. Comments in the file provide the specification. For example, create aMyCustomGenePatternAuthentication.javainterface. - Compile the interface and add it to the classpath for the GenePattern server web application.
- Modify the
authentication.classproperty in the GenePattern configuration file,GenePatternServer/resources/genepattern.properties, to point to the new interface. For example:
authentication.class=org.genepattern.server.auth.MyCustomGenePatternAuthentication - Restart the GenePattern server for the changes to take effect.
See ftp://ftp.broadinstitute.org/pub/genepattern/src/gp-custom-auth.zip for an example project that prepares a custom authentication jar file for deployment to your local GenePattern server.
If the IAuthenticationPlugin interface methods do not provide enough flexibility, you can modify the authentication filter.
Authorization
The authorization filter, AuthorizationFilter.java, controls which GenePattern operations (web pages) the user can access. As described in User Permissions, permissions are based on two configuration files: userGroups.xml, which defines user groups, and permissionMap.xml, which defines which groups have access to which permissions.
Organizations that have user groups defined in an external system can use those groups rather than using the userGroups.xml. To have the authorization filter use external user groups rather than the userGroups.xml file, implement the IGroupMembershipPlugin.java interface:
- Implement the
IGroupMembershipPlugininterface. Use theIGroupMembershipPlugin.javafile as the starting point. Comments in the file provide the specification. For example, create aMyCustomGroupMembershipPlugin.javainterface. - Compile the interface and add it to the classpath for the GenePattern server web application.
- Modify the
group.membership.classproperty in the GenePattern configuration file,GenePatternServer/resources/genepattern.properties, to point to the new interface. For example:
group.membership.class=org.genepattern.server.auth.MyCustomGroupMembershipPlugin - Restart the GenePattern server for the changes to take effect.
To assign permissions to a group authorized through the IGroupMembershipPlugin interface, include the group in the permissionMap.xml file. If the IGroupMembershipPlugin interface methods do not provide enough flexibility, you can modify the authorization filter.
Modifying the filters
The authentication and authorization filters are servlet filters installed in front of the GenePattern web application in the GenePatternServer/Tomcat/webapps/gp/WEB-INF/web.xml file. To implement an alternative authentication (or authorization) filter:
- Write and compile a new
ServletFilterthat that performs the desired authentication (or authorization). - Place the jar file containing the new
ServletFilterinto the following directory:
*/GenePatternServer/Tomcat/webapps/gp/WEB-INF/lib - Modify the GenePattern server’s
web.xmldocument.
*/GenePatternServer/Tomcat/webapps/gp/WEB-INF/web.xml
Note: It is important to maintain the existing order of the servlet filters in theweb.xmldocument as they are used in the order they are defined in the document. The Authentication filter must come before the Authorization filter for the Authorization filter to work. - Change the definition of the
AuthenticationFilter(orAuthorizationFilter) to use the class that you have provided. - Add any necessary configuration elements that it requires.
- Restart the GenePattern server for the changes to take effect.
Note: If you look at the code for the default Authentication Filter (AuthenticationFilter.java), you will see that it allows requests through that have a parameter called jsp_precompile that have come from the localhost. If you do not allow these requests through unauthenticated, you will see a series of errors when you start the GenePattern server as it attempts to precompile the JSP pages. These are not fatal errors, but they slow down server response for users the first time that pages are accessed following a server restart.
Secure Sockets Layer (SSL) Support
This section describes how you can modify the GenePattern web application to run on a web server that is configured to use the HTTPS protocol, where essentially the regular http requests are routed through a secure sockets layer (SSL) making them much harder for hackers to access. If you have installed your GenePattern server onto a web server other than the default Tomcat instance it is distributed with, configure your web server according to its instructions and then follow Step 2 below.
Note: When running under SSL, programming language clients and the GenePattern web client may not be able to connect to your GenePattern server.
Step 1. Configure Tomcat for SSL support
Follow the instructions available at http://tomcat.apache.org/tomcat-5.5-doc/ssl-howto.html to configure the Tomcat instance for using SSL. In doing so, you will modify the Tomcat configuration file, which is located in the GenePatternServer/Tomcat/conf directory.
Step 2.Configure GenePattern for SSL
Once the Tomcat (or other web server) has been configured for SSL, modify the GenePattern configuration file, GenePatternServer/resources/genepattern.properties, to ensure that its properties are in synch with the web server:
- Add a new key,
java.net.ssl.trustStore=<path to keystore>.
This should point to the keystore you created when configuring Tomcat (above) or some keystore that GenePattern can use to establish SSL connections. - Modify the value for the key GENEPATTERN_PORT to use the https port you selected when configuring Tomcat (above).
- Modify the value for the key GenePatternURL to use the https protocol and the https port you selected, for example:
http://localhost:8080/gpbecomeshttps://localhost:8443/gp
Save the genepattern.properties file and restart your server. Any bookmarked links to your GenePattern server must be updated to the new protocol and port.
Changing the GenePattern Database (HSQL to Oracle)
The GenePattern server runs against a database. By default, the GenePattern installation sets up an HSQL database. This section describes how to build and use an Oracle database in place of the HSQL database.
When using Oracle (or another database) you must initialize the database by running the scripts in the <GenePattern_HOME>/resources directory. You or your database administrator must ensure the database is available by JDBC URL from the GenePattern server.
Note: This procedure has been tested using the default Tomcat 5.5 server (Tomcat documentation), which comes with GenePattern.
- Edit the
genepattern.propertiesfiledatabase.vendor=ORACLE# optionally, set a different hibernate.configuration filehibernate.configuration.file=hibernate.oracle.jndi.cfg.xml - Edit
META-INF/context.xml- The
Tomcat/webapps/gp/META-INF/context.xmlfile has several example configurations for connecting the GP server to a database. - Follow the instructions and examples in the comments. Don't forget to set the username and passwords correctly, as well as entering the server name and sid into the URL property. The resource name you use in the
context.xmlfile must match the one you use in the hibernate configuration file.
- The
- Edit the hibernate configuration file
- Edit the
hibernate.configuration.filethat was set in thegenepattern.propertiesfile. This file is loaded relative to the classes directory of your web application. For a default installation the file is here:Tomcat/WEB-INF/classes/hibernate.cfg.xml
- There are some vendor-specific database settings that must be changed from the default when using Oracle. The specific settings needed are in the comments.
- Also be sure to change the
hibernate.connection.datasourceproperty near the top of the file to point to the correct Resource incontext.xml.
- Edit the
- Initialize the database schema
There is a list of files in the installed resources directory for initializing the DB scheme. Run each of theanalysis_oracle-3.X.X.sqlscripts in order, by version number, up through the installed version of GenePattern. - Start your server
Once the GenePattern Server is running, review thegenepattern.logfile to verify that the server started correctly and was able to connect to the database.
Integrating GenePattern with Existing Tools
This section provides guidance to system administrators interested in integrating GenePattern into the analysis tools at their site. It highlights issues that might arise and how to address them, and provides links to relevant portions of the GenePattern documentation, supplementing that documentation as needed.
- Installing GenePattern
- GenePattern Database
- Securing the GenePattern Server
- Modules
- genepattern.properties
Typographical conventions:
|
|
Tables like this describe implementation on the GenePattern public server. |
Installing GenePattern
The standard installation procedure uses Install Anywhere to install the server on Windows, Mac, or Linux using a Tomcat web server. To install on a different web server or on another platform, use the WAR file installer. Instructions for both the standard installation and the WAR file installation are on the download page: http://www.genepattern.org/download/.
Hardware and software requirements for GenePattern are described in the Release Notes.
GenePattern Database
The GenePattern server runs against a database. The GenePattern installation creates an HSQL database. For instructions on how to build and use an Oracle database instead, see Changing the GenePattern Database (HSQL to Oracle).
|
|
We use an Oracle database for the GenePattern public server. |
Securing the GenePattern Server
The following sections briefly summarize how to secure your GenePattern server, including access to the server from client machines, GenePattern user accounts, authentication (e.g., username & password) and authorization (e.g., permissions). For more detail, see Securing the Server.
Access
By default, any client machine can access a GenePattern server. Optionally, you can configure your GenePattern server to restrict access to selected domains. See Securing the Server.Access Filtering.
|
|
Access to the GenePattern public server is not restricted. |
User Accounts
A user must have a GenePattern account to log into the GenePattern server. By default, when a user first logs into the server, GenePattern automatically create an account for that username.
To enable registration, in the genepattern.properties file, set require.password=true. This setting adds a registration link (and password prompt) to the GenePattern login page. The first time users log into GenePattern, they must click the registration link to create an account. User account information is stored in the GenePattern Database.
Alternatively, configure the GenePattern server to not allow users to create GenePattern accounts (create.account.allowed=false). In this case, new user accounts must be explicitly created by editing the GenePattern database.
See Securing the Server.Password Protection.
|
|
Registration (and passwords) are enabled on the GenePattern public server |
Authentication
Each GenePattern user must register to access the GenePattern server. By default, GenePattern requires only a username for authentication. Optionally, you can configure the GenePattern server to require both a username and a password for authentication. See Securing the Server.Password Protection.
GenePattern user authentication is performed by a servlet filter installed in front of the GenePattern web application in its web.xml file. To provide additional or alternative authentication, implement an IAuthenticationPlugin.java interface or modify the servlet filter. See Securing the Server.User Authentication and Authorization.
|
|
The GenePattern public server hosted at the Broad Institute uses the username and password authentication provided by the GenePattern installation. |
|
Collaborator |
A large university uses Kerberos to provide username and password authentication for their network. They wrote their own servlet filter to have the GenePattern server also authenticate using Kerberos. |
Permissions
GenePattern permissions are based on two configuration files:
userGroups.xmldefines user groupspermissionMap.xmldefines which user groups have which permissions; the permissions themselves (e.g., CreateModule, adminModules, and so on) are predefined and cannot be added or removed
GenePattern user authorization is performed by a servlet filter installed in front of the GenePattern web application in its web.xml file. By default, users are assigned permissions based on GenePattern groups. To have the authorization filter use external user groups rather than the userGroups.xml file, implement the IGroupMembershipPlugin.java interface. To provide additional or alternative authorization, modify the servlet filter. See Securing the Server.User Authentication and Authorization.
|
|
On the GenePattern public server, the following permissions are restricted to a small number of users in the Administrator group:
|
SSL
For information on how to modify the GenePattern web application to run on a web server that is configured to use the HTTPS protocol, see Securing the Server.Secure Sockets Layer (SSL) Support.
|
|
The GenePattern public server is not running under SSL. |
Other Security Considerations
We take the following additional steps to secure the machine running the GenePattern public server (these steps may not be necessary on less public servers):
- create an operating system user account with limited permissions from which to run the GenePattern server
- disable JSP compilation and remove the compiler
- prevent users from entering an input file path (file:// urls) as an input file for a module.
By default allow.input.file.paths=false, preventing users from providing an arbitrary network file path (such asfile:///server/directory/file.gct) as the value for an input file parameter. Whenallow.input.file.paths=true, you can use theserver.browse.file.system.rootproperty to set a root directory where the GenePattern server begins browsing for the specified network file path; to do so, editgenepattern.propertiesand setallow.server.file.paths=true - prevent LSF log files from being displayed with other job results in GenePattern; to do so, edit
genepattern.propertiesand setjobs.FilenameFilter=.lsf*(further discussion in Running Modules in a Cluster)
Modules
This section discusses how to install, create, and manage modules.
Installing Modules
By default, you install modules, pipelines, and suites from the Broad repository. The module repository contains more than 100 modules and pipelines. Suites are stored in a separate suite repository. For instructions on how to install modules from the repository, see Managing Modules, Pipelines, and Suites.
The repository is updated regularly. We recommend checking for new modules on a weekly basis.
Create your own repository: Optionally, you can select an alternate repository from which to install modules, pipelines, and/or suites. See Repositories.
|
|
At the Broad, we maintain a development repository for modules in development and a production repository for released modules. Only the production repository is available from the GenePattern public server. |
Creating Modules
For instructions on how to create modules, as well as a step-by-step tutorial for creating a module, see the Programmers Guide.
Running Modules in a Cluster
Queuing systems such as the Load Sharing Facility (LSF) and the Sun Grid Engine (SGE) allow computational resources to be used effectively. If you have such a queuing system, you typically want the GenePattern server to use it. For instructions on how to configure the GenePattern server to use a queuing system, see Using a Queuing System.
As described in the instructions, you click Administration>Server Settings and use the Command Line Prefix page to define the command prefix that runs the module on the cluster. The instructions use the Default Command Prefix field of the Command Line Prefix page to define one command prefix for all modules, which sends all modules to one queue. You can use that same page to define unique command line prefixes for specific modules. This allows you to send different modules to different queues, which helps to address hardware and memory issues. For example, certain modules (such as SNPFileCreator or HierarchicalClustering) require significant amounts of RAM.
The script described in the instructions writes the LSF log file into the job results directory. To prevent GenePattern from displaying the LSF log files with the rest of the job results, edit the genepattern.properties file and set jobs.FilenameFilter=.lsf*.
|
|
The GenePattern public server uses two queues: one for most modules and one for modules that require large amounts of memory. Modules sent to the 'bigmem' queue are run on a cluster of large memory machines. LSF log files are hidden. |
Managing Memory for Modules
In GenePattern, you manage memory for modules in one of two ways:
- Add appropriate parameters to the command line that executes the module. Click Administration>Server Settings and use the Programming Languages page to add memory parameters (e.g., -Xmx512M) for:
- modules written in Java (Java VMOptions field)
- modules written in R (R Options field)
- visualizer modules (Java Visualizer VMOptions field) - Visualizer modules are run on the client machine; therefore, individual users can override this setting by clicking My Settings>Visualizer Memory and specifying the Java Visualizer VMOptions appropriate for their machine.
- Override memory parameters on a per-module basis. Create a file,
java_flags.properties, in the GenePattern/resourcesdirectory. Each line of the file lists the LSID of a module and the memory setting for that module. To find the LSID of a module, in GenePattern, click the module and then click the Properties link. Following is an example file:
# Here is an example which allocates extra RAM for some of the modules: urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00087=-Xmx2500m urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00086=-Xmx10G urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00085=-Xmx2500m urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00106=-Xmx2500m urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00096=-Xmx2500m urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00094=-Xmx2500m urn\:lsid\:broadinstitute.org\:cancer.software.genepattern.module.analysis \:00093=-Xmx2500m
Modules that Require Extra Memory
The following modules frequently require additional memory:
- HierarchicalClustering
- PreprocessDataset
- SNP Analysis modules
- CopyNumberDivideByNormals
- GISTIC
- GLAD
- SNPFileCreator
- SNPFileSorter
- SNPMultipleSampleAnalysis
- XChromosomeCorrect
|
On the GenePattern public server, these modules are sent to a cluster of large memory machines. |
genepattern.properties
Most server configuration options are in the genepattern.properties file in the GenePattern resources directory. Most of the options in this file can be set through the GenePattern interface by clicking Administration>Server Settings. For descriptions of the options, see Modifying Server Settings.
The options listed in the following table can only be set by editing the genepattern.properties settings. We recommend editing the properties through the GenePattern user interface when possible.
|
|
See User Accounts |
|
|
See the FAQ: How do I configure the GenePattern server on a machine with multiple IP addresses? |
|
|
|
|
|
Determines how GenePattern handles network file paths:
|
|
|
Used for the GenePattern SOAP interface. Specify a temporary directory to be used for SOAP messages with attachments. |
Web Service Interface
All GenePattern server functionality is available programmatically. There are two basic access methods:
- Option 1, use one of our programming libraries (Java, MATLAB, or R). The libraries are designed to allow you to use the programming environment as a client, running modules and retrieving results from the GenePattern server. See the Programmers Guide for more information and examples of how to use the libraries.
- Option 2, use SOAP calls directly to http://genepattern.broadinstitute.org/gp/services. This is a standard SOAP interface. The SOAP client interacts with the GenePattern interface as it would with any other SOAP interface.
Setting Up a New GenePattern AMI
The following steps are necessary to create a new GenePattern instance from the GenePattern Amazon Machine Image (AMI).
Create your Amazon account (if you do not already have one)
- Set up an Amazon Web Services account.
- Go to http://aws.amazon.com and click Sign Up Now. Sign in to an existing Amazon account or create a new one.
- Go to http://aws.amazon.com/ec2 and click Sign Up Now. After completing sign up, you should be at the EC2 console.
- In the Navigation menu, click Key Pairs.
- Click the Create Key Pair button and then save the key pair to a file. Name it something you will remember, like "GenePattern." This is private key that is used in creating SSH connections to new AMI instances.
- Set up your Amazon API credentials.
- Select Account>Security Credentials.
- Click the X.509 Certificates tab.
- Click to Create a New Certificate. Note: An Amazon account may have two certificates. If your account already has two certificates, you will need to use one of them, or delete one and create a new one.
- Download the private key and certificate, and save them in a known location, for example. ~/.ec2/prikey.pem and ~/.ec2/cert.pem.
- Make sure these files have the correct security, otherwise using them for SSH will not work: chmod 600 ~/.ec2/*.pem
Create a new instance of the GenePattern AMI
- Go to the AMI creation link: https://console.aws.amazon.com/ec2/home?region=us-east-1#launchAmi=ami-71ba771c
- If you are not already logged in, log into Amazon Web Services.
- If Amazon prompts you for a region, select us-east-1a, as the GenePattern AMI has only been made available for that region.
- Follow the steps in the wizard.
- Make sure to select the Key Pair you just created.
- Feel free to use the default Security Group. This group should have port 8080 open by default. If you are using a custom security group, make sure that port 8080 is open.
- The new instance will appear in your list of instances in the AWS Management Console.
- SSH into the new GenePattern AMI instance using username and password "genepattern".
- Edit /home/genepattern/GenePatternServer/resources/genepattern.properties by changing GenePatternURL to point to the instance URL.
- If you are using your own domain name, you will need to point the domain name at the Amazon Public DNS address. Your domain name provider should have more information on how to do that.
- If you are not using a domain name, the address you will need to enter is the Amazon Public DNS. This can be found by selecting the newly-created GenePattern instance in the EC2 console.
Mounting EBS storage for use with GenePattern (optional)
Note: If one opts not to set up EBS storage, files will be saved on the GenePattern instance's file system. This file system is sufficient only for a small number of GenePattern files.
- Create the EBS
- Go to http://aws.amazon.com/ec2 and sign in.
- Select Volumes>Create Volume.
- When the dialog box appears, enter the size of the EBS volume you want to create. We recommend at least 100 GB for GenePattern. If you are asked for a snapshot, select No Snapshot. This EBS has to be in the same availability zone as the GenePattern instance (us-east-1a).
- Note the ID of the EBS once created. You will need this shortly.
- Attach the EBS to the GenePattern Instance
- Under Volumes, select the EBS volume that you have just created.
- Click Attach Volume and select your GenePattern instance from the list.
- Click Attach and wait several minutes for the volume to finish attaching.
- Note the device name of the attached EBS.
- Mount the EBS in the GenePattern Instance
- SSH into your GenePattern instance using username and password "genepattern".
- Confirm that the EBS has finished attaching by running "cat /proc/partitions" and making sure the attached name appears in the list.
- If you need to format the EBS, run "sudo /sbin/fdisk ATTACHED" where "ATTACHED" is the attached name you were shown for the EBS, for example "/dev/sdk". Follow the prompts. This should give you a partition name, for example "/dev/sdk1". Make note of this.
- If you need to partition the EBS (you will need to partition the EBS unless you are using a pre-existing EBS), run "sudo /sbin/mkfs -t ext3 PARTITION" where "PARTITION" is the partition name you were just given.
- To mount the partition, run "sudo mount PARTITION /home/genepattern/GenePatternServer/data" where "PARTITION" is the partition name you were just given.
Start your GenePattern AMI
- You can start GenePattern by running: /home/genepattern/GenePatternServer/StartGenePatternServer
- You can view your GenePattern instance by navigating to http://ADDRESS:8080/gp where ADDRESS is the domain name in use or the Amazon Public DNS address.
- GenePattern log files can be found at: /home/genepattern/GenePatternServer/Tomcat/logs/
Running GenePattern in a Docker container
Since the GenePattern server (as of 2017) primarily uses Docker to encapsulate the runtime environments for its modules, Docker is required to run any modules of recent vintage. Therefore Docker is essential on a GenePattern server host whether or not the server itself is run inside a Docker container. For information about Docker itself, see the official docker website for documentation and tutorials. The following section details considerations and steps used to run GenePattern in Docker on a Macintosh OSX computer. For running in Docker on other operating systems, similar issues and approaches should be viable.
When running the GenePattern server in Docker, or with Docker modules, you should consider the security issues involved. When modules are sent to the Docker demon on the host, the module containers could potentially write files to any parts of the host disk drives that have been mounted to them as the root user. In the case of Genepattern, this typically means the TaskLib, Users and JobResults directories. For this reason many IT organizations do not like to allow Docker on internal servers. This can be mitigated by using Singularity instead of Docker for the module images which has been done in practice, but as of this writing (May 2020) we have not yet developed and documented a simple automated system for doing so. This security risk will be mitigated in the future as it is possible to both run the Docker demon under a user account, and to set up networking without privileged mode, when running Docker on Linux. We will update these instructions once that is available on Macintosh OSX.
Running modules on the host Docker demon has the advantage that each module runs in its own seperate container allowing better utilization of compute resources and, if a docker swarm or other distributed docker system is in play, module executions can easily be distributed to multiple computers or virtual machines.
Windows10 Setup Instructions
For Windows10, installation of docker requires WSL (Windows Subsystem for Linux) version 2 as described in the Docker documentation. To use the start-genepattern.sh script described below, you will also need to make sure that your default WSL distribution is also version 2. To check run "wsl -l -v".
e.g.
C:\Users\liefe> wsl -l -v
NAME STATE VERSION
* Ubuntu Running 2
docker-desktop-data Running 2
docker-desktop Running 2
If your default is using version 1, you can change it using the following 2 commands. This assumes your default distribution is Ubuntu.
wsl set-version Ubuntu 2
wsl set-default-version 2
Once this is done you can start wsl at a Powershell prompt
wsl
and follow the instructions for linux/MacOS platforms below.
Note: The "start-genepattern.sh" script (see below) will not run properly in Powershell due to the different slashes used for file system paths between Windows and Linux. You must either start wsl first and execute it there, or use "bash .\start-genepattern.sh" at a Powershell prompt which will use bash within WSL to execute the script rather than Powershell itself.
Overview of running GenePattern Server in Docker
When running GenePattern Server from a docker container, it is assumed that this is desired to be a persistent GenePattern installation. Therefore the details of jobs run and analysis output files are kept external to the docker container. This allows them to remain accessible even if the server container is deleted, and to be connected to updated versions of the container as they become available. The directories that are kep external are
- users - where uploaded user files are kept
- jobResults - where job output files are kept
- taskLib - where module code for legacy (pre-docker) modules is kept
- resources - where configuration files are kept. Also holds the GenePattern database if the default HSQL database is used.
Latest versions of scripts and example configuration files referenced below are available at https://github.com/genepattern/genepattern-server/tree/develop/docker
When running the GenePattern server in docker, the following configuration changes are made (relative to the default installation using the installer);
- "<genepattern_home>/resources/wrapper_scripts/run-with-docker.sh" is set as the default command-line prefix.
- "job.docker.bind_src" is set to your local <genepattern_home> directory. This directory (and its sub directories) will be writable to the GenePattern server and module containers.
- "job.docker.bind_dst" is set to "/opt/genepattern". This is the mounting point for your "<genepattern_home>" directory within the module containers.
Steps for running GenePattern in docker on a host computer
The following steps were developed on a Macintosh OSX laptop. The scripts should work on most Linux installations with the bash shell installed, but the scripts can be customized as needed for other environments. We recommend that Windows users run the GenePattern server container from WSL*
- Decide on the GenePattern server container version you wish to run. GenePattern server containers are publicly available in dockerhub and use a versioning system that matches the github releases for the project.
Copy or make note of the version tag you have selected, e.g. v3.9_21.04.05_b325
- Create a root server directory for the GenePattern installation. All GenePattern files and folders will be created in or below this directory.
- Copy the start-genepattern.sh script from the GenePattern github repository into your root server directory.
- Run the start-genepattern.sh script. This script will pull the Docker image, create the directories, customize the config_custom.yaml for the server, and create a docker container with the necessary port mappings and volume mounts. The desired GenePattern server tag should be passed in with the -v or --version flag.
- e.g.
./start-genepattern.sh -v v3.9_21.04.05_b325
The container name will be "genepattern" but can be editted by changing the variable at the top of the script.
- Login to the GenePattern server at http://127.0.0.1:8080/gp and complete the installation. You will need to register your server and then install modules etc as per a non-Docker installation.
Following installation, the server can be stopped via the usual docker command (e.g. "docker stop genepattern"). Restarting the container can be done via re-running the start-genepattern.sh script or via the docker start command. When re-running the start-genepattern.sh script, it will check for a running container or a pre-existing but stopped GenePattern container rather than creating an installation.
*If you choose to run GenePattern in Docker from Windows (not using WSL) you will need to alter the Docker run command. The steps to do so are as follows:
- Run steps 1-4 as above (NB your server will not start)
- copy the
docker runcommand from the second to the last line of the start-genepattern.sh script. - In a terminal or notepad...etc replace all instances of $PWD in the run command with the Unix version of your root server directory for GenePattern. E.G. If you created a root server directory C:\Users\Me\gp_home the Unix version would be /c/Users/Me/gp_home
- Replace
$namewith "genepattern" and$VERSIONwith the version of the GenePattern server Docker container you created when you first ran the start-genepattern.sh script - it will look like this:docker run -v /c/Users/Me/gp_home:/c/Users/Me/gp_home -v /var/run/docker.sock:/var/run/docker.sock -w /c/Users/Me/gp_home -v /c/Users/Me/gp_home/resources:/opt/genepattern/resources -v /c/Users/Me/gp_home/taskLib:/opt/genepattern/taskLib -v /c/Users/Me/gp_home/jobResults:/opt/genepattern/jobResults -v /c/Users/Me/gp_home/users:/opt/genepattern/users -p 8888:8888 -p 8080:8080 -d --name "genepattern" genepattern/genepattern-server:v3.9_21.07_b352 /opt/genepattern/StartGenePatternServer
- Run that command in the terminal (preferably PowerShell) and then navigate to 127.0.0.1:8080/gp in a browser window.
Changing the server version container
When you wish to upgrade the version of the GenePattern server container, you will want to save your users, uploaded files, job results and configuration. This is also built into the start-genepattern.sh script which will preserve and reuse the following files (if run in the same directory as the previous version).
- "users" directory
- "jobResults" directory
- "tasklib" directory
- resources/genepattern.properties
- resources/database_custom.properties
- resources/config_custom.yaml
- resources/userGroups.xml
- resources/PermissionsMap.xml
- resources/GenepatternDB.properties
- resources/GenePatternDB.script
The process to update the Genepattern server to a newer version of the container is as follows;
- Stop the running server
docker stop genepattern
- Remove the old container
docker rm genepattern
- Start a new container with the disired version using the start-genepattern.sh script and desired new tag (where the new tag replaces the v3.9_21.04.05_b325 in the following command
start-genepattern.sh -v v3.9_21.04.05_b325
- Login to the GenePattern server at http://127.0.0.1:8080/gp and commence using it.
Integrating GenePattern with Globus
See how to use Globus in GenePattern in the User Guide.
One option for getting data to a GenePattern server is to integrate it with a Globus endpoint and to use the Globus network to transfer data betweeen other Globus endpoints, and the GenePattern server. When operating in this manner, GenePattern will use a mapped collection on the endpoint. Since GenePattern accounts do not map to the underlying unix system, the configuration of the Globus collection and GenePattern server is not typical for Globus setups.
In broad strokes, the integration looks like this:
- A Globus endpoint is created, if necessary. On that endpoint a guest collection is created.
- The GenePattern server to connect is registered with Globus auth.
- When transfers are to be made
- GenePattern creates an directory for the user on the guest collection.
- GenePattern creates an ACL for the Globus user on the guest collection directory, permitting read or write as needed.
- GenePattern initates the transfer on behalf of the user
- GenePattern removes the ACL after the transfer completes
- GenePattern moves the transferred files out of the guest collection and into GenePatterns working directories
The reason for the guest collection and ACL creation/destruction is that the number of ACL's is limited to no more than a few thousand, while GenePattern has tens of thousands of users. Therefore we cannot assign permanent ACL entries.
Create a Globus endpoint and guest collection
See the Globus documentation for creating an endpoint at https://www.globus.org/globus-connect.
See the Globus documentation for creating a guest collection at https://docs.globus.org/how-to/create-collections-posix/#:~:text=Click%20the%20Shares%20tab%20and,Click%20Allow.
Register the GenePattern server with Globus auth
See the globus developer documentation at https://docs.globus.org/api/auth/developer-guide/#:~:text=Globus%20requires%20client%20applications%20(apps,app%20registration%20responsibilities%20to%20others.
Configure GenePattern to use Globus
Here is an example configuration for a GenePattern server that will use a globus connect endpoint on the same host machine.
###############################
#
# Globus OAuth config settings
#
##############################
oauth.authorize.url: "https://auth.globus.org/v2/oauth2/authorize"
oauth.client.id: "xxxxxxxx-yyyy-zzzz-aaaa-bbbbbbbbbbbb"
oauth.client.secret: "your client secret here"
oauth.client.scopes: "urn:globus:auth:scope:transfer.api.globus.org:all urn:globus:auth:scope:auth.globus.org:view_identities openid profile email offline_access"
authentication.class: org.genepattern.server.webapp.rest.api.v1.oauth.GlobusAuthentication
In this configuration example, the following configuration items are required.
oauth.authorize.url: This is always "https://auth.globus.org/v2/oauth2/authorize"
oauth.client.id: This is the client ID created when your GenePattern server was registered with Globus
oauth.client.secret: This is the client ID created when your GenePattern server was registered with Globus.
oauth.client.scopes: These are always "urn:globus:auth:scope:transfer.api.globus.org:all urn:globus:auth:scope:auth.globus.org:view_identities openid profile email offline_access" and reflect the details the GenePattern server needs to allow login through Globus, set up the ACLs and transfer files in Globus on behalf of the users.
authentication.class: This is always "org.genepattern.server.webapp.rest.api.v1.oauth.GlobusAuthentication" and tells the GenePattern server that it must allow Globus for authentication as well as the normal GenePattern authentication. Note that users need accounts both with GenePattern and with Globus to use this. Users may create new GenePattern accounts by logging into GenePattern through Globus with a Globus Identity, or they can map a Globus identity to a GenePattern account in the GenePattern User Profile pages.
In addition you need to tell GenePattern where your endpoint is, the type of endpoint (local or S3) and where to find the files. For a local Globus Personal Connect endpoint, you need these three additional configuration parameters.
# # globus connect endpoint using a globus connect personal endpoint
###############
globus.local.endpoint.id: "xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxx"
globus.local.endpoint.root: "/Users/myusername/GlobusEndpoint/GenePatternLocal/"
globus.local.endpoint.type: "localfile"
globus.local.endpoint.id: This is the Globus endpoint ID as can be found in https://app.globus.org
globus.local.endpoint.root: This is the path to the guest collection in the file system. In the example it is "/Users/myusername/GlobusEndpoint/GenePatternLocal/" but it should match the path to the guest collection on your Globus Connect endpoint guest collection. The user running the GenePattern server must have read/write permissions on this directory.
globus.local.endpoint.type: For a Globus endpoint on the local file system this should be "localfile". For an example using an S3 backed endpoint see below.
For an S3 backed endpoint, the additional parameters are slightly different. Note that an S3 backed endpoint can only be used on a GenePattern server that uses "org.genepattern.server.executor.awsbatch.AWSS3ExternalFileManager" as the external file manager class.
# root = path on the endpoint, root.s3 is the actual S3 path of the endpoint
###############
globus.local.endpoint.id: "zzzzzzzz-zzzz-zzzz-zzzz-zzzzzzzzzzzz"
globus.local.endpoint.root: "/tedslaptop/"
globus.local.endpoint.root.s3: "s3://MyBucketName/GlobusConnectSharedRoot/GenePatternLocal/"
globus.local.endpoint.type: "S3"
globus.local.endpoint.id: This is the Globus endpoint ID as can be found in https://app.globus.org
globus.local.endpoint.root.s3: This defines the S3 url and directory path that is used for the guest collection of the S3 backed endpoint. In the example the bucket is named "MyBucketName" and should be changed to match a bucket you own or have read/write access to. The path to the (virtual) directory on S3 in the example is "GlobusConnectSharedRoot/GenePatternLocal/" and should be changed to what ever path you want to use as a key prefix for objects transferred in from Globus.
globus.local.endpoint.type: For an S3 backed endpoint, this should simply be "S3".
