Overview
The GenePattern flow cytometry suite was developed by the Brinkman lab at the British Columbia Cancer Research Center (BCCRC), who collaborated with the GenePattern team to make these modules publicly available and easily accessible in GenePattern. The suite consists of 34 modules in 3 general categories, discussed below.
The workflow associated with these modules (as described in the forthcoming article) is outlined here:
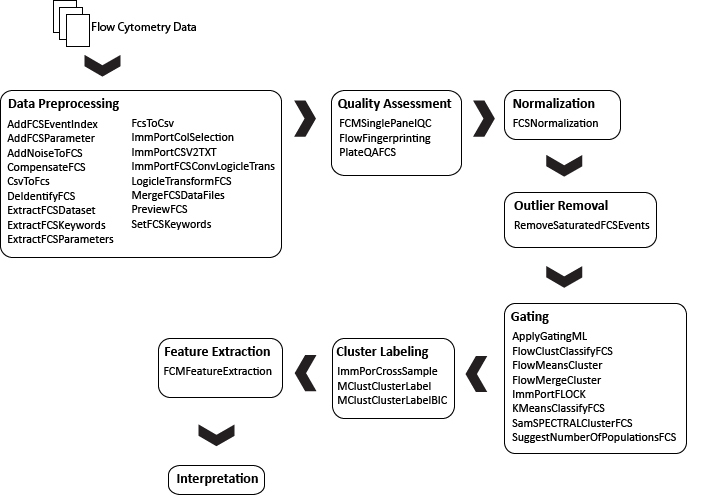
Note: The ImmPort modules are designed to work together to process data files and perform density-based clustering for automated population identification of multi-dimensional flow cytometry data based on the FLOCK (FLOw cytometry Clustering without K) algorithm. For more information about these modules, read their associated documentation. In addition, there are two pipelines to facilitate use of the ImmPort modules.
Categories Within the Suite
There are three categories of modules within the suite, covering data preprocessing, quality assessment, and gating and clustering. The modules in these suites may span multiple steps of the flow cytometry flowchart.
Data Preprocessing
Flow cytometry Data Preprocessing suite includes various utilities for FCS data preview and transformations, conversion between spreadsheets (i.e., CSV files) FCS files, merging and sub-sampling data, editing keywords in FCS files and other support tools.
Quality Assessment
Flow cytometry Quality Assessment suite includes modules implementing various approaches to automatically assess the quality of flow cytometry data. Such an assessment represents an important part of any data analysis, and quality control tests should be included at the beginning of data analysis and often at other steps of an analytical pipeline to identify differences in samples originating from changes in conditions that are probably not biologically motivated. Generally, these methods establish a quality control criterion to give special consideration to abnormal samples or even exclude these from further analysis.
Gating and Clustering
Flow cytometry Gating and Clustering suite includes modules for the application of manually created gates (saved in Gating-ML) on FCS data files and various clustering algorithms developed for the use with flow cytometry data.
References
This is the complete list of references for all the modules in the suite.
- FCS: Spidlen J, Moore W, Parks D, Goldberg M, Bray C, Bierre P, Gorombey P, Hyun B, Hubbard M, Lange S, Lefebvre R, Leif RR, Novo D, Ostruszka L, Treister A, Wood J, Murphy RF, Roederer M, Sudar D, Zigon R, Brinkman RR. Data File Standard for Flow Cytometry, Version FCS 3.1. Cytometry A. 2010;77:97–100.
- FlowClust: Lo K, Brinkman RR, Gottardo R. Automated gating of flow cytometry data via robust model-based clustering. Cytometry A. 2008;73(4):321-332.
- FlowMeans: Aghaeepour N, Nikolic R, Hoos HH, Brinkman RR. Rapid cell population identification in flow cytometry data. Cytometry A. 2011;79(1):6-13.
- FlowMerge: Finak G, Bashashati A, Brinkman R, Gottardo R. Merging mixture components for cell population identification in flow cytometry. Adv Bioinformatics. 2009;247646. Epub 2009 Nov 12.
- FlowQ: Le Meur N, Rossini A, Gasparetto M, Smith C, Brinkman RR, Gentleman R. Data quality assessment of ungated flow cytometry data in high throughput experiments. Cytometry A. 2007;71(6):393-403.
- Fingerprinting: Rogers WT, Holyst H.FlowFP: A bioconductor package for fingerprinting flow cytometric data. Adv Bioinformatics. 2009:193947. Epub 2009 Sep 24.
- Gating-ML: Spidlen J, Leif RC, Moore W, Roederer M, International Society for the Advancement of Cytometry Data Standards Task Force, Brinkman RR. Gating-ML: XML-based gating descriptions in flow cytometry. Cytometry A. 2008;73A(12):1151-1157.
- ImmPortFCSConvLogicleTrans (FCSTrans) Qian Y, Liu Y, Campbell J, Thomson E, Kong YM, Scheuermann, RH. FCSTrans: An open source software system for FCS file conversion and data transformation. Cytometry A. 2012:81;353-356. doi: 10.1002/cyto.a.22037
- ImmPortFLOCK: Qian Y, Wei C, Eun-Hyung Lee F, Campbell J, Halliley J, Lee JA, Cai J, Kong YM, Sadat E, Thomson E, Dunn P, Seegmiller AC, Karandikar NJ, Tipton CM, Mosmann T, Sanz I, Scheuermann RH. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin Cytom. 2010;78 Suppl 1:S69-82.
- Logicle: Moore W, Parks D. Update for the logicle data scale including operational code implementations. Cytometry A. 2012;81(4):273-277.
- Normalization: Hahne F, Khodabakhshi AH, Bashashati A, Wong C-J, Gascoyne RD, Weng AP, Seifert-Margolis V, Bourcier K, Asare A, Lumley T, Gentleman R, Brinkman RR. Per-channel basis normalization methods for flow cytometry data. Cytometry A. 2010;77(2):121-131.
- SamSPECTRAL: Zare H, Shooshtari P, Gupta A, Brinkman RR. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics. 2010 Jul 28;11:403.
